Abstract
Disruption of DNA methylation patterns is one of the hallmarks of cancer. Similar to other cancer types, human papillomavirus (HPV)-driven head and neck cancer (HNC) also reveals alterations in its methylation profile. The intrinsic ability of HPV oncoproteins E6 and E7 to interfere with DNA methyltransferase activity contributes to these methylation changes. There are many genes that have been reported to be differentially methylated in HPV-driven HNC. Some of these genes are involved in major cellular pathways, indicating that DNA methylation, at least in certain instances, may contribute to the development and progression of HPV-driven HNC. Furthermore, the HPV genome itself becomes a target of the cellular DNA methylation machinery. Some of these methylation changes appearing in the viral long control region (LCR) may contribute to uncontrolled oncoprotein expression, leading to carcinogenesis. Consistent with these observations, demethylation therapy appears to have significant effects on HPV-driven HNC. This review article comprehensively summarizes DNA methylation changes and their diagnostic and therapeutic indications in HPV-driven HNC.
1. HPV-Driven Head and Neck Cancer
Head and neck cancers (HNCs) are a diverse group of cancers arising in the upper aerodigestive tract, which includes the oral cavity, pharynx, larynx, nasal cavity, and paranasal sinuses [1]. Conventionally, the global burden of HNC has been mainly attributed to tobacco exposure and excessive alcohol consumption [2]. However, during the last two decades, the epidemiological landscape of HNC has been changed considerably, especially in developed countries, due to the continuous increase in human papillomavirus (HPV)-driven HNC [3,4,5]. Despite the available prophylactic vaccination program, a significant reduction in HPV-driven HNC incidence is not expected until 2060, necessitating further investigations to improve disease prevention, diagnosis, and treatment [3].
The oropharynx can be considered as the hotspot of HPV-driven cancers in the head and neck region, as the vast majority of these cancers arise in the palatine tonsils and base of the tongue [6,7]. These cancers have clinically and biologically distinct features from their HPV-negative counterparts in terms of their gene expression profiles, mutation burden, epigenetic profiles, and treatment response [8,9,10,11]. Despite the aggressive nature of HPV-driven HNCs, which often metastasize into nearby lymph nodes, these cancers are associated with a favourable prognosis, as they respond well to treatment [1]. However, the underlying biology of these cancers associated with these distinct features is poorly understood, especially the association between tumor attributes and epigenetic alterations.
Disruption of normal methylation patterns is one of the hallmarks of cancer. These changes are often associated with the tumorigenic process and tumor characteristics, such as tumor progression and metastasis [12]. However, HPV-driven cancers are unique, as viral proteins also contribute to these methylation changes. Studies conducted to interrogate the methylation landscape of HPV-driven HNC have revealed many inherent features. This review aims to summarise and evaluate the current state of knowledge on DNA methylation changes associated with HPV-driven HNCs and to discuss the diagnostic and therapeutic implications of these methylation changes.
2. Human Papillomavirus
Papillomaviruses are non-enveloped, icosahedral viruses, which belong to the Papillomaviridae family [13]. Over 400 papillomaviruses have been identified to date, and among them there are more than 200 types of papillomaviruses that can infect humans [14,15]. Papillomaviruses are epitheliotropic viruses, as their life cycles evolved to be dependent on the differentiation process of the epithelial tissue. Most of the papillomaviruses cause chronic asymptomatic infection with minimum effect on the host. However, several types (around 20) have evolved sophisticated mechanisms to promote cellular transformation [16]. These high-risk types are well known for their association with anogenital cancers and the increasing incidence of HNC. Among all HPV types, type 16 is associated with the majority of HPV-driven cancers, irrespective of the site [5].
The oncogenic potential of HPV is mainly attributed to HPV oncoproteins E5, E6, and E7 [17]. Through promoting cell proliferation and masking of the cell from the host’s surveillance mechanisms, these proteins allow cells to accumulate genetic and epigenetic changes over time, leading to oncogenesis [18]. These are multipurpose proteins that can interact and manipulate many cellular pathways [18]. Among the interactions, E7 protein’s ability to bind with retinoblastoma protein in order to hinder sequestration of E2F transcription factors, leading to transcriptional activation; and E6 protein’s ability to downregulate p53 in order to avoid cell cycle arrest and apoptosis are considered to play central roles [19].
Moreover, HPV oncoproteins E6 and E7 are also known for their ability to interact with cellular epigenetic machinery [20]. These interactions further promote genetic and epigenetic instability, and facilitate oncogenesis and tumor progression. Studies conducted to investigate epigenetic changes in HNC have detected many changes appearing in the epigenome. Among them, DNA methylation is one of the most studied epigenetic markers due to its possible utility in disease identification and treatment interventions. Early diagnosis of HNC has been one of the major concerns of the studies performed to date. Furthermore, DNA methylation has been identified to be involved in tumorigenesis, tumor progression, and metastasis across many types of cancer, rendering it a potential avenue for treatment interventions [21].
3. DNA Methylation
The covalent modification of bases in the DNA strand by the addition of methyl groups is referred to as DNA methylation. In eukaryotes, DNA methylation primarily involves cytosine [22,23]. In higher-order animals such as vertebrates, DNA methylation, when present, almost exclusively presents in the 5’ position of the cytosine ring [10,22,24]. Furthermore, cytosine methylation predominantly appears in cytosine bases, which are positioned 5’ to guanines (CpG dinucleotides) in the DNA strand [25,26].
Cytosine methylation is known to play a crucial role in genome stabilization. It is involved in gene imprinting, X chromosome inactivation, and repression of transposons and retroviral sequences in the genome [25]. In synchronization with histone modification, cytosine methylation orchestrates gene expression [26]. In most of the genes, regulatory sequences, such as promoters and enhancers, overlap with CpG islands [23,27]. These are the genes that are vulnerable to methylation-associated gene regulation [23]. Under normal conditions, most of the active and inactive genes have unmethylated regulatory sequences, indicating that DNA methylation is not the sole system of annotating genetic information [25]. However, it is consistently observed that cytosine methylation involving CpG islands in the regulatory sequences leads to transcriptional silencing of genes, and active genes often have hypomethylated or unmethylated regulatory sequences [23,28].
DNA methyltransferases (DNMT) are the enzymes that catalyze DNA methylation and are involved in both the establishment and maintenance of DNA methylation. DNMT1 is responsible for the transfer of methylation patterns to the newly synthesized DNA strand following DNA replication [29]. DNMT3A and DNMT3B, on the other hand, are involved in de novo methylation [30,31]. However, it has been shown that DNMT3A and DNMT3B are also required for methylation establishment and maintenance in addition to DNAMT1 [32]. In contrast, ten eleven translocation (TET) proteins counter the activity of DNMTs. TET1, TET2, and TET3 are capable of demethylating cytosines through a series of reactions initiating from oxidizing 5-methylcytosine to 5-hydroxymethylcytosine (5hmC) [33,34]. Moreover, studies indicate that activation-induced cytidine deaminase (AICDA) and thymine DNA glycosylase (TDG) are also involved in the demethylation process [35,36].
At the functional level, interpretation of the established epigenetic annotation involves many protein factors, such as the methyl-CpG-binding domain (MBD) family of proteins [26,37]. These factors, when bound to methylated DNA, are capable of restricting the access for transcription factors causing downregulation of the gene expression [38]. Furthermore, members of MBD family proteins are capable of interacting with histone modification enzymes, promoting a more closed chromatin structure in order to restrict the access for the transcription machinery [39,40].
4. DNA Methylation and Cancer
Disruption of DNA methylation patterns is often observed in cancer. In many cancer types, it has been observed that the genomes of cancer cells undergo an overall decrease in the level of 5-methylcytosines, while having increased 5-methylcytosines in the regulatory regions [41,42]. In certain instances, these changes, resulting in the inactivation of tumor suppressor genes and activation of oncogenes, act as alternates or complementary mechanisms to gene alterations, promoting carcinogenesis [43,44]. Moreover, certain methylation changes may enhance tumor progression and aggressiveness by promoting cell proliferation, disruption of cellular communication, promoting cell migration, downregulation of apoptosis, etc. [43,45].
5. DNA Methylation in HPV-Driven HNC
Similar to other cancer types, HPV-driven HNCs also reveal alterations in their methylation profiles [20,46]. However, due to the fact that HPV oncoproteins E6 and E7 are capable of interfering with cellular DNA methylation machinery (Figure 1), DNA methylation becomes more intricate, thus complicating interpretation in HPV-driven cancers [20].
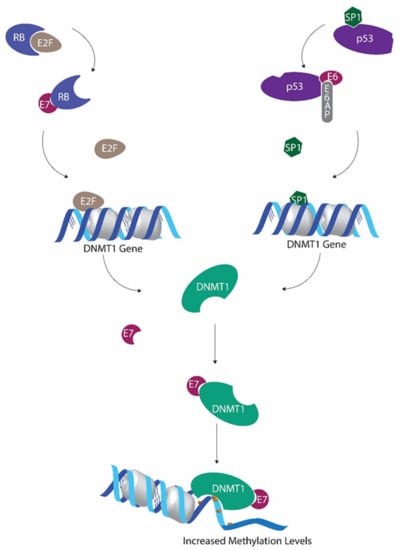
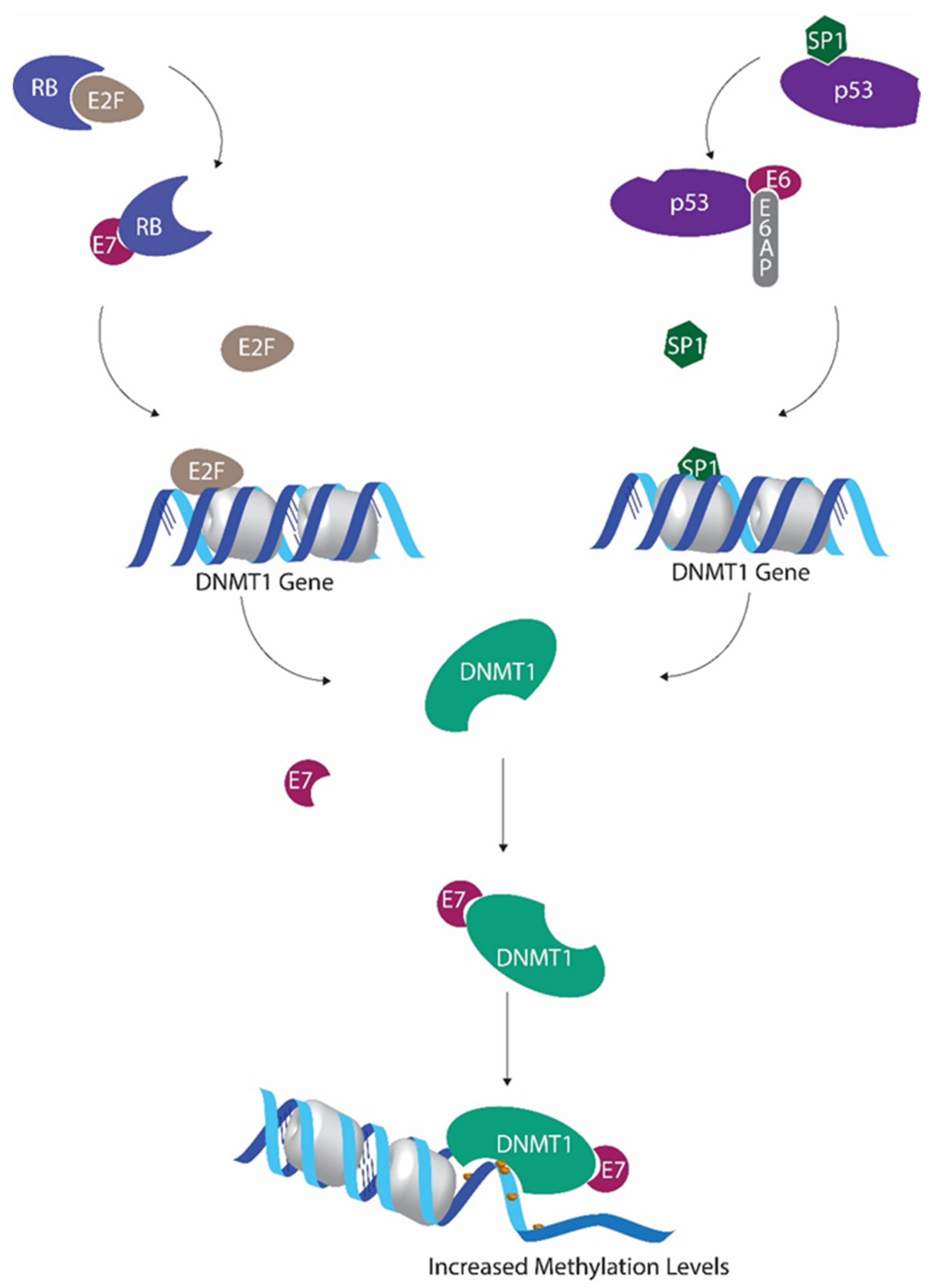
Figure 1.
Human papillomavirus (HPV) E6 and E7 mediated regulation of methyltransferase activity. P53 inactivation by E6 leads to decreased Sp1 transcriptional factor inactivation, allowing increased DNMT1 transcription. E7-mediated downregulation of retinoblastoma protein hinders E2F sequestration. This E2F upsurge further promotes DNMT1 expression. E7 binds with DNMT1 to form the E7/DNMT1 complex, promoting DNMT1 activity.
It has been shown that global cytosine methylation levels are altered when p53 is inactivated [47,48]. Wild-type p53 is capable of binding and repressing the DNMT1 promoter in cooperation with specificity protein 1 (Sp1) [47]. As a result, when p53 is deactivated, DNMT1 levels tend to increase [47]. Suppression of p53 by HPV E6 is one of the main effects of HPV activity. In addition to all the other detrimental outcomes of p53 inactivation, this also results in DNMT1 upregulation [49].
The ability of HPV E7 protein to promote E2F activity by downregulating retinoblastoma protein (pRb) also promotes DNMT1 transcription, as E2F transcription factors have the ability to activate DNMT1 promoter [50,51]. Furthermore, HPV E7 also has the ability to bind with DNMT1 using the CR3 zinc finger domain located in the C terminal and to enhance the activity of DNMT1 directly [20].
6. Alterations in Methylation Patterns in HPV-Driven HNC
Studies conducted to investigate methylation patterns of HPV-driven HNC have demonstrated the ability of HPV oncoproteins to promote DNMT activity. Many of these studies have also revealed a significant increase in methylation levels, especially in the promoter regions, in HPV-driven HNC [46,52]. Moreover, transfection studies have confirmed the ability of HPV oncoproteins to promote DNA methylation [53]. However, certain genes have also been reported to be hypomethylated in HPV-driven HNC compared to non-HPV-driven HNC (Table 1).

Table 1.
Differentially methylated genes and loci in HPV-driven head and neck cancer (HNC).
7. Commonly Reported Differentially Methylated Genes in HPV-Driven HNC
7.1. Genes Involved in Cell Cycle Regulation and Programmed Cell Death
7.1.1. CDKN2A
Changes in methylation patterns of the CDKN2A gene have been reported in several studies. This gene, positioned at 9p21.3, encodes two regulatory proteins, namely p16INK4a and p14ARF [76]. Both these proteins function as cell cycle regulators and are considered as important tumor suppressors; p14 ARF is involved in stabilization of p53 by downregulating MDM2 mediated p53 ubiquitination [77], while p16INK4a is a cyclin-dependent kinase (CDK) inhibitor involved in the retinoblastoma (pRb) pathway [78]. Genetic and epigenetic alterations in the CDKN2A gene resulting in expression changes have been observed in many types of cancer [79,80,81,82]. However, CDKN2A expression is considered even more imperative in HPV-driven HNC, as p16INK4a expression itself is considered as an independent prognosticator of HPV-driven oropharyngeal cancer (OPC) [83]. HPV-driven cancers usually present with higher p16INK4a levels attributable to E7-mediated repression of the pRB pathway [19,84]. Thus, in HPV-driven OPC, the CDKN2A gene is expected to be active and to have hypomethylated or unmethylated regulatory sequences.
Several studies have reported lower methylation levels in the CDKN2A locus in HPV-driven HNC compared to non-HPV-driven HNC [11,65,70]. This finding is not only seen in tumor tissue; hypomethylation in CKND2A has also been reported in salivary samples of HPV-positive HNC patients compared to healthy controls [58].
In contrast to these findings, several other studies have suggested that the gene is hypermethylated in HPV-driven HNCs compared to their non-HPV-driven counterparts. Schlecht et al. (2015) reported hypermethylation in non-promoter CpG islands in the CDKN2A gene compared to adjacent normal mucosa [1]. Swangphon et al. (2017) and Choudhury et al. (2015) reported CDKN2A promoter hypermethylation in HPV-positive HNC compared to normal control tissue [56,60]. Similarly, cervical cancer studies and penile cancer studies have reported hypermethylation in the CKDN2A locus compared to precancer [85,86,87,88,89]. Some studies have also reported CDKN2A hypermethylation in HPV-driven HNC in subjects with decreased P16 expression [56,64]. A similar observation has also been reported in a penile cancer study [89]. Despite these findings, some other studies that have been conducted to investigate CKDN2A methylation in HPV-driven HNC have not detected any significant methylation changes, indicating the heterogeneity of CKDN2A methylation patterns in HPV-driven cancers [67,72].
7.1.2. RASSF1
The Ras association domain family 1 (RASSF1) gene located in the 3p21.3 position encodes for several protein isoforms containing a Ras association domain [90]. Among these isoforms, RASSF1A is considered to have tumor suppressor properties and has been shown to be silenced by genetic and epigenetic mechanisms in several types of cancer [90,91]. The RASSF1 gene has been reported to be hypermethylated in HPV-related cancers, such as cervical cancer and penile cancer [88,92,93]. Consistent with these observations, Choudhury et al. (2015) reported promoter hypermethylation in the RASSF1 gene in HPV-driven HNCs compared to non-HPV-driven HNCs [60]. However, this contradicts the findings of Colacino et al. (2013) and Dong et al. (2003), who reported hypomethylation of this gene in HPV-driven HNCs compared to their non-HPV-driven counterparts [65,75].
7.1.3. CCNA1
The CCNA1 gene at 13q13.3 encodes for cyclin A1 protein. Even though this protein is not the major A-type cyclin involved in the cell cycle, it has been shown in murine models that its absence slows down the cell cycle in somatic cells [94,95]. Furthermore, together with CDK2, cyclin A1 has been shown to regulate double-strand break repair in DNA [96]. Promoter hypermethylation in CCNA1 gene has been reported in HPV-driven HNCs compared to non-HPV-driven HNCs in several studies [11,65,67]. Furthermore, promoter hypermethylation of the gene has also been reported in several other cancers, such as cervical cancer and nasopharyngeal carcinoma [97,98,99,100].
7.2. Genes Involved in Cellular Adhesion and Communication
7.2.1. Cadherin Family Genes
The cadherin family of proteins is considered as the major class of cellular adhesion molecules, which are known to play crucial roles in cell signalling and communication, especially in solid tissues [101]. Proper cellular adhesion and communication ensure cellular coordination, which is critical to normal cellular differentiation, tissue morphogenesis, and homeostasis. The weakening of cellular adhesion associated with genetic and epigenetic changes in cadherin genes is often observed in epithelial cancers [102]. Methylation changes in cadherin genes have been reported in several HNC studies. Promoter hypermethylation of the CDH8 gene located at 16q21, encoding a type II cadherin, is reported in several studies [46,53,59]. Promoter methylation of the same gene has also been reported in cervical cancer [103]. Furthermore, promoter hypermethylation of several other cadherin genes, including CDH 18, CDH15, CDH13, CDH19, and CDH23, has been reported by Lechner et al. (2013) in HPV-driven HNC [53]. Hypermethylation in the CDH11 locus also has been reported in HPV-driven HNC in a different study [65]. Similar methylation patterns in cadherin genes, such as CDH1 and CDH13, have been reported in cervical cancer [104,105,106,107].
Moreover, promoter hypermethylation of several protocadherin genes has also been reported in the literature. Among them, the protocadherin beta 11 (PCDHB11) locus has been reported to be hypermethylated in HPV-driven HNC compared to non-HPV-driven HNC [53,59]. Furthermore, hypermethylation in several other protocadherin genes, namely PCDH8, PCDH9, PCDHB3, PCDH10, PCDH15, PCDHB1, PCDHB4, and PCDHB15, has also been reported in HPV-driven HNC (53). When other HPV-associated cancers are considered, promoter methylation in PCDHA4, PCDH10, and PCDHA 13 has been reported in cervical cancer [108,109].
7.2.2. ITGA4
Integrin alpha 4 (ITGA4) gene has also been reported to be hypermethylated in HPV-driven HNC in a few studies (46, 55). ITGA4 gene has also been reported to be hypermethylated in cervical cancer [110]. Many other non-HPV-driven cancer studies have reported similar findings [111,112]. The gene positioned at 2q31.3 encodes for alpha 4 peptides, which function as crucial integrins in association with beta 1 or beta 7 peptides [113,114]. These integrins mediate cellular adhesion, migration, and signalling [115]. Alpha 4 integrins are involved in the activation and function of hemopoiesis and leucocyte trafficking, playing an essential role in immunity [116]. Furthermore, alpha 4 integrins have also shown to be involved in the immune response against viral infections [117]. If the hypermethylation reported in the methylation studies is gene silencing in nature, it may have contributed to diminished viral clearance, leading to prolonged HPV infection. However, further studies are necessary to elucidate the existing association.
7.3. Genes Involved in Cellular Migration and Tumor Progression
7.3.1. TIMP3
Matrix metalloproteinases (MMPs), one of the families of natural endopeptidases, are capable of degrading extracellular matrix components and are known to play a significant role in tumor progression [118]. Tissue inhibitor of metalloproteinase (TIMP) acts as the natural inhibitor of MMPs and is known to play a role as a tumor suppressor [119,120,121]. Promoter methylation of the TIMP3 gene located at 22q12.3 has been reported in a few studies in HPV-driven HNCs compared to non-HPV-driven HNCs [52,67].
7.3.2. ELMO1
Engulfment and cell motility 1 protein (ELMO1), encoded by the ELMO1 gene located at 7p14.2-p14.1, is known to promote cellular migration [122]. This has been linked to increased invasion and metastasis in several types of cancer [122,123]. However, methylation studies report the gene to be hypermethylated in HPV-driven HNC [55,59]. It has not been clearly reported whether these hypermethylated CpGs are located in the promoter region of the gene. However, promoter methylation in the gene has been reported in cervical cancer [124].
7.4. Genes and Non-Coding Regions with Unknown Association with Carcinogenesis
7.4.1. MEI1
Lechner et al. (2013) reported the promoter of the MEI1 located at 22q13.2 to be hypomethylated in HPV-driven HNCs compared to non-HPV-driven HNCs. Even though exact sites of methylation have not been specified, hypomethylation in the same locus has been reported Worsham et al. in HPV-driven HNC [59]. The gene encodes for meiotic double-stranded break formation protein 1, which is known to involve in meiosis [125]. A functional association between the gene and cancer has not been reported as of yet.
7.4.2. LINE1
Long interspersed nuclear element 1 (LINE1) is one of the most abundant types of retrotransposons accumulated in the human genome, which accounts for approximately 17% of the total DNA [126]. Most copies of LINE1 have been inactivated by truncations, rearrangements, and mutations, and only several copies retain their ability for retrotransposition [126,127]. DNA methylation plays a key role in silencing these competent LINE1 sequences [128]. Hypermethylation of LINE1 region, in comparison to HPV-negative HNC, has been reported in HPV-driven HNC [11,68,69]. However, LINE1 methylation in HPV-driven HNC compared to healthy controls is not clear, as it has been reported that HPV-negative tumors tend to have hypomethylated LINE1 regions [69].
8. HPV DNA Methylation in HPV-Driven HNC
The involvement of DNA methylation in gene imprinting, X chromosome inactivation, and repression of transposons and retroviral sequences indicates that DNA methylation fundamentally intends to silence the undesirable or harmful sections of the genome in order to achieve genomic stabilization [25]. Possibly for the same purpose, HPV genomes within the host cells become targets of the cellular methylation machinery.
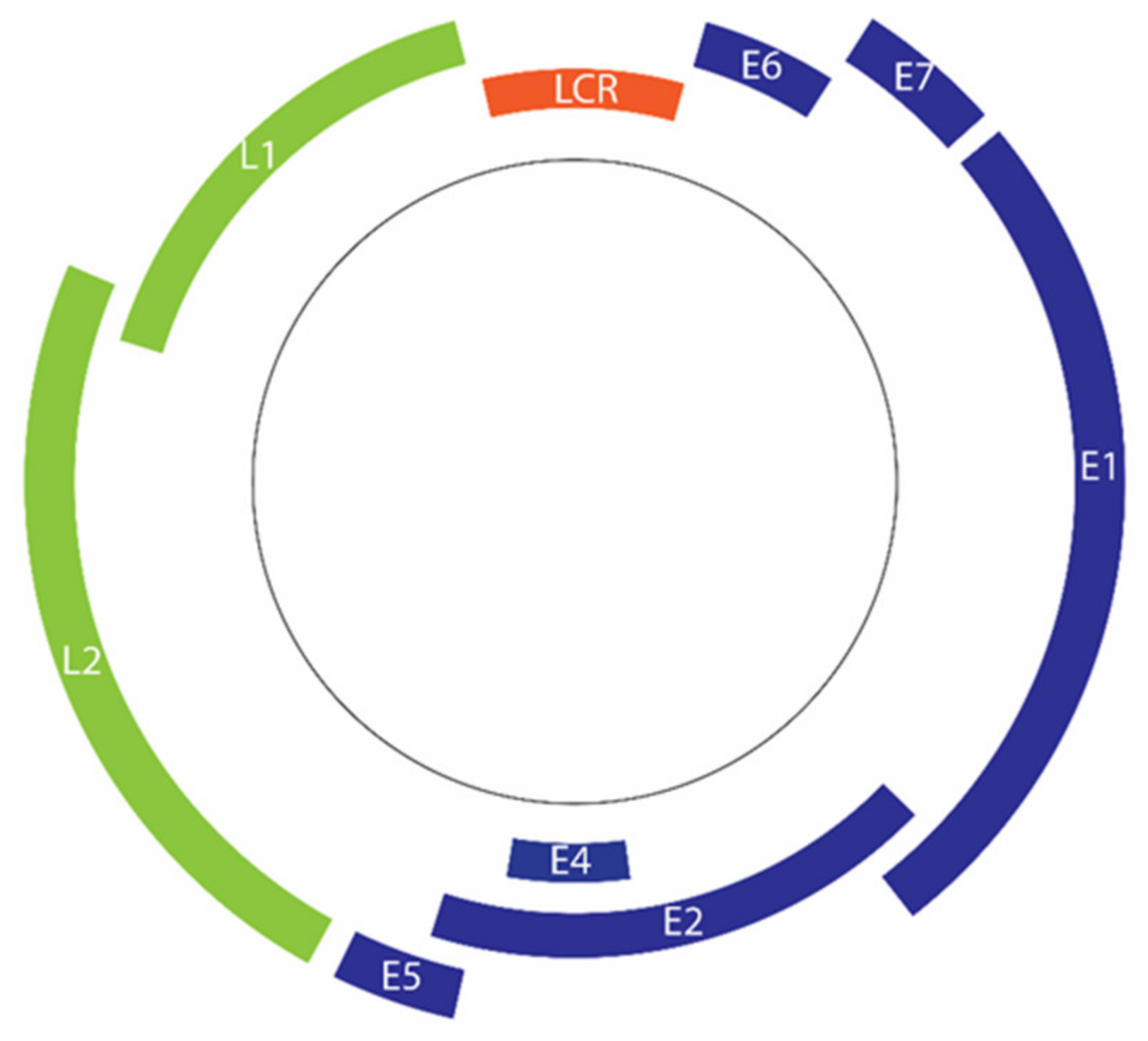
The double-stranded DNA genome of HPV comprises approximately 8000 base pairs [13]. The genome can be divided into 3 regions, namely the early region (E), late region (L), and upstream regulatory region (URR) or long control region (LCR) (Figure 2) [15,129]. The HPV genome has only two common promoters, an early promoter and a later promoter [130]. The early promoter, which is located in the viral LCR, is responsible for the transcription regulation of HPV early proteins, including major oncoproteins E6 and E7 [130,131].
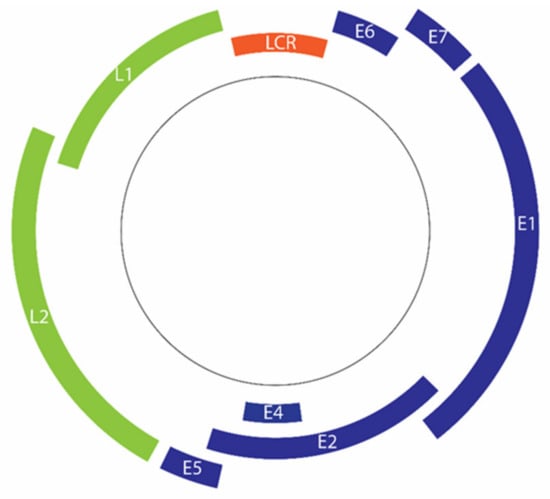
Figure 2.
Organization of the HPV 16 genome. Deferent regions code for early genes (blue) and late genes (green). Transcription of early genes is regulated by the long control region (LCR) (Orange).
Similar to promoter methylation in the host genes, DNA methylation in the viral LCR may downregulate the transcription of early viral genes [132]. This downregulation is often associated with the deregulation of transcription factor binding [132,133,134]. Binding of methyl CpG binding proteins to the methylated sites is also known to obstruct transcription factor binding [132]. However, LCR methylation does not necessarily cause transcriptional silencing. Instead, selective methylation can even cause overexpression of early viral proteins [135].
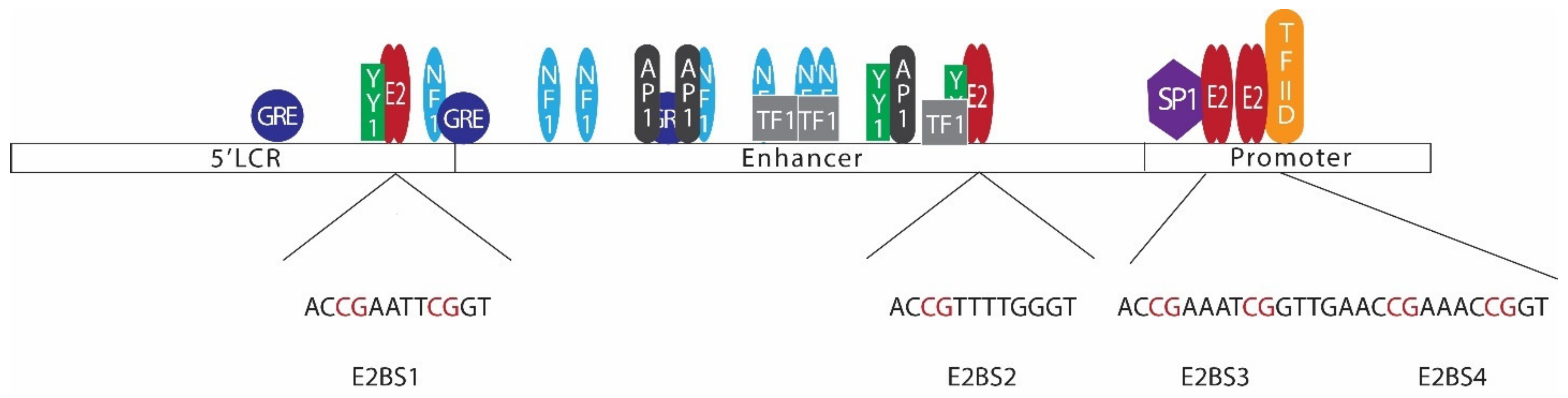
E2, the major regulatory protein of HPV, is capable of controlling the transcription of HPV early proteins through a mechanism involving negative feedback control [136,137]. In most mucosal HPVs, LCRs contain 4 E2 binding sites (Figure 3) [138]. Among those binding sites, E2BS1 located towards the 5’ end of the LCR has the highest affinity for the E2 protein. E2BS1 recruits E2, even at lower concentrations, and this binding activates the viral early promoter, enhancing the transcription of HPV early proteins, including E6, E7, and E2 itself [135,138,139]. As a result, E2 levels increase and E2 starts to bind the other E2 binding sites towards the 3’ end of the viral LCR, which have comparatively lower affinities to E2 [135,138]. This binding, especially with E2BS3 and E2BS4, represses the early promoter, hindering the transcription of early proteins [135]. E2BS3 and E2BS4 are located in close proximity to SP1 and transcription factor II D (TFIID) binding sites, and binding of E2 to these sites displaces SP1 or TFIID, repressing the transcription [134].
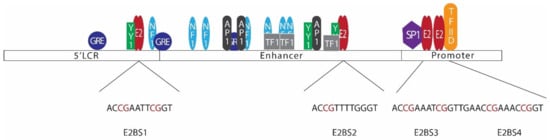
Figure 3.
Arrangement of E2 binding sites in the HPV 16 long control region (LCR). The HPV 16 LCR can be subdivided into 3 sections, namely the 5’LCR, enhancer, and promoter regions. Binding of transcription factors such as YY1, NF1, GRE, AP1, SP1, TF1, and TFIID regulates viral early gene transcription. LCR consists of 4 E2 binding sites with a common palindromic sequence of ACCG(N)4CGGT (E2BS2 has a slight difference in the sequence). Several CpG sites are located in LCR E2 binding sites [138,140,141,142,143].
Alterations in E2 binding sites or E2 protein itself may disrupt this regulation mechanism. For instance, changes in E2BS3 or E2BS4 disrupting the E2 binding to these sites would result in overexpression of HPV early proteins. HPV E2 binding sites have a common palindromic sequence of ACCG(N)4CGGT [135]. E2 binds to these DNA motifs in the form of a dimer, with the DNA binding domain positioned at the carboxy-terminal of the protein [144]. However, CpG methylation in the binding sites hinder E2 binding to the site, disturbing E2 mediated viral early promoter regulation [145]. As such, methylation in E2BS3 or E2BS4 may lead to disruption of E2-mediated transcriptional regulation, leading to overexpression of viral oncoproteins, increasing the odds of tumorigenesis [131,146]. Studies conducted to investigate LCR CpG methylation in HNC have reported varying degrees of methylation levels (Table 2) [147,148,149]. These findings suggest that CpG methylation in LCR is not common to all HPV-driven HNC. Similar findings have also been reported in cervical cancer studies [150,151,152]. LCR methylation, especially CpG sites at E2BS3 and E2BS4, seems to be an alternative pathway of losing E2 control, leading to oncoprotein overexpression. Thus, it is not necessarily present in all the HPV-driven cancers.

Table 2.
HPV DNA methylation in HPV-driven HNC.
Not only in viral LCR, CpG methylation has also been observed in other CpG sites of the HPV genome. In cervical cancer, viral L1 and L2 genes have been reported to have higher methylation levels in cancers compared to low-grade precancer [153,154]. HPV DNA methylation in these sites has not been comprehensively investigated in HPV-driven HNC. However, available literature suggests similar methylation patterns in HNC [54,155]. However, a functional relationship between CpG methylation of these sites and tumorigenesis has not been identified to date. It can be speculated that this type of methylation may result from long term HPV infection with ineffective replication of the late viral genes.
9. Diagnostic Implications
Even though studies have proposed methylation signatures characteristic to HPV-driven HNC, these signatures are not consistent among studies. Except for a few genes, most of the genes reported to be differentially methylated have been reported only in a single study. Thus, the methylation profiles of HPV-driven HNCs appear to be considerably heterogeneous. However, extrapolating the results from different studies and drawing conclusions are challenging issues, as methodologies of the studies are considerably dissimilar in terms of identification of the HPV status of the tumor, evaluation of the methylation status, and the comparisons used.
The majority of studies have reported on methylation status by comparing HPV-driven and non-HPV-driven HNCs, an approach that does not allow identification of methylation changes from normal methylation patterns. Furthermore, these comparisons may erroneously identify hypermethylation in one group as hypomethylation in the other group. Despite these challenges, several genes and loci have been identified to be differentially methylated by independent studies. However, the available information is not sufficient to make firm conclusions. As such, more comprehensive multicentered studies with uniform methodologies are necessary to identify diagnostic methylation targets.
The diagnostic value of HPV DNA methylation patterns also needs to be further investigated. Cervical cancer studies have suggested that methylation changes in HPV L1 and L2 genes can be used to discriminate cervical cancer from low-grade precancer [153,154]. Even though HNC studies have also reported similar findings, these associations have not been comprehensively investigated [54].
The majority of the studies have considered only HPV16, as it is the most common type involved in HPV-driven cancers, irrespective of the site. Studies that have considered other high risk (HR-HPV) types have not reported separate data for these HR-HPV types, possibly due to the lower number of samples. Even though these findings are generalized for HR-HPV, characteristic features may exist in cancers caused by different HPV types. However, such interpretations are not possible due to the paucity of available data.
10. Therapeutic Implications
Despite the differences between studies in terms of their methodologies many studies have shown that HPV-driven HNCs have higher overall methylation levels compared to their HPV-negative equivalents [11,46,52,53]. Methylation changes have been reported in genes that are known to be associated with tumorigenesis, tumor progression, and metastasis across many cancer types. Furthermore, methylation changes in the HPV genome leading to oncoprotein overexpression have also been identified. Presumably, these methylation changes may play a role in HPV-driven HNCs, at least in certain instances.
Consistent with these findings, demethylation therapy has shown to be effective against HPV-driven HNCs. In both HPV-positive HNCs and cervical cell lines, demethylating agent 5-aza-2′-deoxycytidine (DAC) has shown efficacy in downregulating HPV E6 and E7 expression [156]. In addition to hindering oncoprotein expression, demethylation treatment has been found to increase p53 and p21 protein levels and tumor suppressor microRNA 375 levels, resulting in an overall decrease in cancer cell growth and survival [156]. Similar observations have been reported in a different study using demethylation agent 5-azacytidine (5-aza) [157]. In vivo studies have confirmed in vitro findings, with similar responses for demethylation therapy by HPV-driven HNC. A study conducted using xenografted tumors in nude mice reported tumor growth inhibition [157]. The same study also reported a marked decrease in HPV oncoprotein expression, p53 stabilization, and upregulated caspase activity in HPV-driven HNC patients [157].
Furthermore, an increased interferon response in certain HPV-driven HNC cell lines and a decrease in matrix metalloproteinases (MMP) expression in both mouse models and HPV-driven HNC patients following demethylation therapy were reported in the same study [157]. There is a possibility that these changes are associated with the activation of host genes following demethylation therapy. For instance, it has been reported that promoter methylation of the TIMP3 gene, which encodes a natural metalloproteinase, is commonly seen in HPV-driven HNC [52,67]. Activation of the TIMP3 gene following demethylation therapy would result in an increase in the TIMP3 level, eventually inhibiting MMP endopeptidases.
Several clinical trials have been conducted to evaluate the effectiveness of demethylation therapy on HNC [158]. A specific clinical trial to assess the effectiveness of demethylation therapy on HPV-driven HNC is currently underway (NCT02178072).
11. Conclusions
Many changes in the host’s methylation landscape have been observed in HPV-driven HNC. However, the proposed methylation signatures are not consistent among studies; except for a few genes and loci, the vast majority have not been repeatedly reported in studies. Despite these inconsistencies, many studies have reported overall higher methylation levels in HPV-driven HNC. Furthermore, several genes that are known to be associated with tumorigenesis, tumor progression, and metastasis have been reported to be differentially methylated in HPV-driven HNC. In addition to methylation changes in the host genome, methylation changes in the HPV genome have also been reported. Consistent with these findings, demethylation therapy has been shown to have significant effects on HPV-driven HNC cancer.
Author Contributions
Conceptualization, C.P., K.D.T., and C.E.W.; data curation, C.P., K.D.T.; writing—original draft preparation, C.E.W.; writing—review and editing, K.D.T., S.V., J.L.-L., L.K., and C.P.; visualization, C.E.W.; supervision, K.D.T., S.V., J.L.-L., L.K., and C.P.; funding acquisition, C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Cancer Australia (APP1145657) and the Garnett Passé and Rodney Williams Foundation funding for Chamindie Punyadeera.
Acknowledgments
Ekanayake W.S. Chameera was supported by a scholarship from the University Grants Commission, Sri Lanka, and Queensland University of Technology, Australia
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Chaturvedi, A.K.; Anderson, W.F.; Fakhry, C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 3235–3242. [Google Scholar] [CrossRef] [PubMed]
- Berman, T.A.; Schiller, J.T. Human papillomavirus in cervical cancer and oropharyngeal cancer: One cause, two diseases. Cancer 2017, 123, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Liederbach, E.; Kyrillos, A.; Wang, C.H.; Liu, J.C.; Sturgis, E.M.; Bhayani, M.K. The national landscape of human papillomavirus-associated oropharynx squamous cell carcinoma. Int. J. Cancer 2017, 140, 504–512. [Google Scholar] [CrossRef]
- Tang, K.D.; Baeten, K.; Kenny, L.; Frazer, I.H.; Scheper, G.; Punyadeera, C. Unlocking the Potential of Saliva-Based Test to Detect HPV-16-Driven Oropharyngeal Cancer. Cancers 2019, 11, 473. [Google Scholar] [CrossRef]
- Lohavanichbutr, P.; Houck, J.; Fan, W.; Yueh, B.; Mendez, E.; Futran, N.; Doody, D.R.; Upton, M.P.; Farwell, D.G.; Schwartz, S.M.; et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: Potential implications for treatment choices. Arch. Otolaryngol. Head Neck Surg. 2009, 135, 180–188. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Furlan, C.; Lupato, V.; Polesel, J.; Fratta, E. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: Role of HPV and lifestyle factors. Clin. Epigenet. 2017, 9, 124. [Google Scholar] [CrossRef]
- Sartor, M.A.; Dolinoy, D.C.; Jones, T.R.; Colacino, J.A.; Prince, M.E.; Carey, T.E.; Rozek, L.S. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 2011, 6, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Rodger, E.J.; Eccles, M.R. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol. 2018, 51, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. S1), 2–23. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Munger, K. Expert Views on HPV Infection. Viruses 2018, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Araldi, R.P.; Assaf, S.M.R.; Carvalho, R.F.; Carvalho, M.; Souza, J.M.; Magnelli, R.F.; Modolo, D.G.; Roperto, F.P.; Stocco, R.C.; Becak, W. Papillomaviruses: A systematic review. Genet. Mol. Biol. 2017, 40, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Tommasino, M.; Depuydt, C.; Dillner, J. Are 20 human papillomavirus types causing cervical cancer? J. Pathol. 2014, 234, 431–435. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Estevao, D.; Costa, N.R.; Gil da Costa, R.M.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef]
- Burgers, W.A.; Blanchon, L.; Pradhan, S.; de Launoit, Y.; Kouzarides, T.; Fuks, F. Viral oncoproteins target the DNA methyltransferases. Oncogene 2007, 26, 1650–1655. [Google Scholar] [CrossRef]
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA Methylation in Cancer and Aging. Cancer Res. 2016, 76, 3446–3450. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Romero, M.A.; Cota, I.; Casadesus, J. DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 2015, 25, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N. Engl. J. Med. 2018, 378, 1323–1334. [Google Scholar] [CrossRef]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Bell, J.S.K.; Vertino, P.M. Orphan CpG islands define a novel class of highly active enhancers. Epigenetics 2017, 12, 449–464. [Google Scholar] [CrossRef]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005, 2 (Suppl. S1), S4–S11. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Baubec, T.; Colombo, D.F.; Wirbelauer, C.; Schmidt, J.; Burger, L.; Krebs, A.R.; Akalin, A.; Schubeler, D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 2015, 520, 243–247. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Liang, G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009, 10, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, Z.; Cheng, J.; Rao, Q.; Gong, W.; Liu, M.; Shi, Y.G.; Zhu, J.; Wang, P.; Xu, Y. Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell 2013, 155, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Lu, J.; Cheng, J.; Rao, Q.; Li, Z.; Hou, H.; Lou, Z.; Zhang, L.; Li, W.; Gong, W.; et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 2015, 527, 118–122. [Google Scholar] [CrossRef]
- Cortellino, S.; Xu, J.; Sannai, M.; Moore, R.; Caretti, E.; Cigliano, A.; Le Coz, M.; Devarajan, K.; Wessels, A.; Soprano, D.; et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011, 146, 67–79. [Google Scholar] [CrossRef]
- Bhutani, N.; Brady, J.J.; Damian, M.; Sacco, A.; Corbel, S.Y.; Blau, H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 2010, 463, 1042–1047. [Google Scholar] [CrossRef]
- Domcke, S.; Bardet, A.F.; Adrian Ginno, P.; Hartl, D.; Burger, L.; Schubeler, D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015, 528, 575–579. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Stirzaker, C.; Song, J.Z.; Ng, W.; Du, Q.; Armstrong, N.J.; Locke, W.J.; Statham, A.L.; French, H.; Pidsley, R.; Valdes-Mora, F.; et al. Methyl-CpG-binding protein MBD2 plays a key role in maintenance and spread of DNA methylation at CpG islands and shores in cancer. Oncogene 2017, 36, 1328–1338. [Google Scholar] [CrossRef]
- Cramer, J.M.; Pohlmann, D.; Gomez, F.; Mark, L.; Kornegay, B.; Hall, C.; Siraliev-Perez, E.; Walavalkar, N.M.; Sperlazza, M.J.; Bilinovich, S.; et al. Methylation specific targeting of a chromatin remodeling complex from sponges to humans. Sci. Rep. 2017, 7, 40674. [Google Scholar] [CrossRef]
- Sina, A.A.I.; Carrascosa, L.G.; Liang, Z.; Grewal, Y.S.; Wardiana, A.; Shiddiky, M.J.A.; Gardiner, R.A.; Samaratunga, H.; Gandhi, M.K.; Scott, R.J.; et al. Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat. Commun. 2018, 9, 4915. [Google Scholar] [CrossRef]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Shlomi, T.; Rabinowitz, J.D. Metabolism: Cancer mistunes methylation. Nat. Chem. Biol. 2013, 9, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef] [PubMed]
- Van Tongelen, A.; Loriot, A.; De Smet, C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017, 396, 130–137. [Google Scholar] [CrossRef]
- Nakagawa, T.; Matsusaka, K.; Misawa, K.; Ota, S.; Takane, K.; Fukuyo, M.; Rahmutulla, B.; Shinohara, K.I.; Kunii, N.; Sakurai, D.; et al. Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett. 2017, 407, 21–31. [Google Scholar] [CrossRef]
- Lin, R.K.; Wu, C.Y.; Chang, J.W.; Juan, L.J.; Hsu, H.S.; Chen, C.Y.; Lu, Y.Y.; Tang, Y.A.; Yang, Y.C.; Yang, P.C.; et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010, 70, 5807–5817. [Google Scholar] [CrossRef]
- Peterson, E.J.; Bogler, O.; Taylor, S.M. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res. 2003, 63, 6579–6582. [Google Scholar]
- Au Yeung, C.L.; Tsang, W.P.; Tsang, T.Y.; Co, N.N.; Yau, P.L.; Kwok, T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010, 24, 1599–1604. [Google Scholar]
- McCabe, M.T.; Davis, J.N.; Day, M.L. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005, 65, 3624–3632. [Google Scholar] [CrossRef]
- Leonard, S.M.; Wei, W.; Collins, S.I.; Pereira, M.; Diyaf, A.; Constandinou-Williams, C.; Young, L.S.; Roberts, S.; Woodman, C.B. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis 2012, 33, 1286–1293. [Google Scholar] [CrossRef]
- Van Kempen, P.M.; van Bockel, L.; Braunius, W.W.; Moelans, C.B.; van Olst, M.; de Jong, R.; Stegeman, I.; van Diest, P.J.; Grolman, W.; Willems, S.M. HPV-positive oropharyngeal squamous cell carcinoma is associated with TIMP3 and CADM1 promoter hypermethylation. Cancer Med. 2014, 3, 1185–1196. [Google Scholar] [CrossRef]
- Lechner, M.; Fenton, T.; West, J.; Wilson, G.; Feber, A.; Henderson, S.; Thirlwell, C.; Dibra, H.K.; Jay, A.; Butcher, L.; et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013, 5, 15. [Google Scholar] [CrossRef]
- Giuliano, A.R.; Nedjai, B.; Lorincz, A.T.; Schell, M.J.; Rahman, S.; Banwait, R.; Boulware, D.; Sirak, B.; Martin-Gomez, L.; Abrahamsen, M.; et al. Methylation of HPV 16 and EPB41L3 in oral gargles: Associations with oropharyngeal cancer detection and tumor characteristics. Int. J. Cancer 2020, 146, 1018–1030. [Google Scholar] [CrossRef]
- Ren, S.; Gaykalova, D.; Wang, J.; Guo, T.; Danilova, L.; Favorov, A.; Fertig, E.; Bishop, J.; Khan, Z.; Flam, E.; et al. Discovery and development of differentially methylated regions in human papillomavirus-related oropharyngeal squamous cell carcinoma. Int. J. Cancer 2018, 143, 2425–2436. [Google Scholar] [CrossRef]
- Swangphon, P.; Pientong, C.; Burassakarn, A.; Vatanasapt, P.; Kleebkaow, P.; Patarapadungkit, N.; Treebupachatsakul, T.; Promthet, S.; Kongyingyoes, B.; Ekalaksananan, T. Methylation Status of P16Ink4a in Human Papillomavirus-Associated Cancer of Oral Cavity and Oropharynx in Northeastern Thailand. Asian Pac. J. Cancer Prev. 2017, 18, 699–705. [Google Scholar] [PubMed]
- Degli Esposti, D.; Sklias, A.; Lima, S.C.; Beghelli-de la Forest Divonne, S.; Cahais, V.; Fernandez-Jimenez, N.; Cros, M.P.; Ecsedi, S.; Cuenin, C.; Bouaoun, L.; et al. Unique DNA methylation signature in HPV-positive head and neck squamous cell carcinomas. Genome Med. 2017, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Wan, Y.; Vagenas, D.; Ovchinnikov, D.A.; Perry, C.F.; Davis, M.J.; Punyadeera, C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 2016, 16, 749. [Google Scholar] [CrossRef]
- Worsham, M.J.; Chen, K.M.; Datta, I.; Stephen, J.K.; Chitale, D.; Gothard, A.; Divine, G. The biological significance of methylome differences in human papilloma virus associated head and neck cancer. Oncol. Lett. 2016, 12, 4949–4956. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, J.H.; Ghosh, S.K. Promoter Hypermethylation Profiling Identifies Subtypes of Head and Neck Cancer with Distinct Viral, Environmental, Genetic and Survival Characteristics. PLoS ONE 2015, 10, e0129808. [Google Scholar] [CrossRef]
- Schlecht, N.F.; Ben-Dayan, M.; Anayannis, N.; Lleras, R.A.; Thomas, C.; Wang, Y.; Smith, R.V.; Burk, R.D.; Harris, T.M.; Childs, G.; et al. Epigenetic changes in the CDKN2A locus are associated with differential expression of P16INK4A and P14ARF in HPV-positive oropharyngeal squamous cell carcinoma. Cancer Med. 2015, 4, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.M.; Stephen, J.K.; Havard, S.; Mahan, M.; Divine, G.; Worsham, M.J. IGSF4 methylation as an independent marker of human papillomavirus-positive oropharyngeal squamous cell carcinoma. JAMA Otolaryngol. Head Neck Surg. 2015, 141, 257–263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kostareli, E.; Holzinger, D.; Bogatyrova, O.; Hielscher, T.; Wichmann, G.; Keck, M.; Lahrmann, B.; Grabe, N.; Flechtenmacher, C.; Schmidt, C.R.; et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J. Clin. Investig. 2013, 123, 2488–2501. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Stockmann, C.; Schrodter, K.; Rudack, C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell. Oncol. 2013, 36, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Colacino, J.A.; Dolinoy, D.C.; Duffy, S.A.; Sartor, M.A.; Chepeha, D.B.; Bradford, C.R.; McHugh, J.B.; Patel, D.A.; Virani, S.; Walline, H.M.; et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS ONE 2013, 8, e54742. [Google Scholar] [CrossRef] [PubMed]
- Gubanova, E.; Brown, B.; Ivanov, S.V.; Helleday, T.; Mills, G.B.; Yarbrough, W.G.; Issaeva, N. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin. Cancer Res. 2012, 18, 1257–1267. [Google Scholar] [CrossRef]
- Weiss, D.; Basel, T.; Sachse, F.; Braeuninger, A.; Rudack, C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol. Carcinog. 2011, 50, 680–688. [Google Scholar] [CrossRef]
- Poage, G.M.; Houseman, E.A.; Christensen, B.C.; Butler, R.A.; Avissar-Whiting, M.; McClean, M.D.; Waterboer, T.; Pawlita, M.; Marsit, C.J.; Kelsey, K.T. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin. Cancer Res. 2011, 17, 3579–3589. [Google Scholar] [CrossRef]
- Richards, K.L.; Zhang, B.; Baggerly, K.A.; Colella, S.; Lang, J.C.; Schuller, D.E.; Krahe, R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS ONE 2009, 4, e4941. [Google Scholar] [CrossRef]
- Taioli, E.; Ragin, C.; Wang, X.H.; Chen, J.; Langevin, S.M.; Brown, A.R.; Gollin, S.M.; Garte, S.; Sobol, R.W. Recurrence in oral and pharyngeal cancer is associated with quantitative MGMT promoter methylation. BMC Cancer 2009, 9, 354. [Google Scholar] [CrossRef]
- Marsit, C.J.; Posner, M.R.; McClean, M.D.; Kelsey, K.T. Hypermethylation of E-cadherin is an independent predictor of improved survival in head and neck squamous cell carcinoma. Cancer 2008, 113, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, E.M.; Toner, M.E.; Finn, S.P.; Fan, C.Y.; Ring, M.; Hagmar, B.; Timon, C.; Smyth, P.; Cahill, S.; Flavin, R.; et al. p16(INK4A) genetic and epigenetic profiles differ in relation to age and site in head and neck squamous cell carcinomas. Hum. Pathol. 2008, 39, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Furniss, C.S.; Marsit, C.J.; Houseman, E.A.; Eddy, K.; Kelsey, K.T. Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Marsit, C.J.; McClean, M.D.; Furniss, C.S.; Kelsey, K.T. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int. J. Cancer 2006, 119, 1761–1766. [Google Scholar] [CrossRef]
- Dong, S.M.; Sun, D.I.; Benoit, N.E.; Kuzmin, I.; Lerman, M.I.; Sidransky, D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin. Cancer Res. 2003, 9 Pt 1, 3635–3640. [Google Scholar]
- Irelan, J.T.; Gutierrez Del Arroyo, A.; Gutierrez, A.; Peters, G.; Quon, K.C.; Miraglia, L.; Chanda, S.K. A functional screen for regulators of CKDN2A reveals MEOX2 as a transcriptional activator of INK4a. PLoS ONE 2009, 4, e5067. [Google Scholar] [CrossRef]
- Bates, S.; Phillips, A.C.; Clark, P.A.; Stott, F.; Peters, G.; Ludwig, R.L.; Vousden, K.H. p14ARF links the tumour suppressors RB and p53. Nature 1998, 395, 124–125. [Google Scholar] [CrossRef]
- Steiner, M.S.; Wang, Y.; Zhang, Y.; Zhang, X.; Lu, Y. p16/MTS1/INK4A suppresses prostate cancer by both pRb dependent and independent pathways. Oncogene 2000, 19, 1297–1306. [Google Scholar] [CrossRef]
- Bisio, A.; Nasti, S.; Jordan, J.J.; Gargiulo, S.; Pastorino, L.; Provenzani, A.; Quattrone, A.; Queirolo, P.; Bianchi-Scarra, G.; Ghiorzo, P.; et al. Functional analysis of CDKN2A/p16INK4a 5’-UTR variants predisposing to melanoma. Hum. Mol. Genet. 2010, 19, 1479–1491. [Google Scholar] [CrossRef]
- Collado, M.; Blasco, M.A.; Serrano, M. Cellular senescence in cancer and aging. Cell 2007, 130, 223–233. [Google Scholar] [CrossRef]
- Brown, V.L.; Harwood, C.A.; Crook, T.; Cronin, J.G.; Kelsell, D.P.; Proby, C.M. p16INK4a and p14ARF tumor suppressor genes are commonly inactivated in cutaneous squamous cell carcinoma. J. Investig. Dermatol. 2004, 122, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.; Sibinga Mulder, B.G.; Bonsing, B.; Morreau, H.; Farina Sarasqueta, A.; Inderson, A.; Luelmo, S.; Feshtali, S.; Potjer, T.P.; de Vos Tot Nederveen Cappel, W.; et al. Risk of multiple pancreatic cancers in CDKN2A-p16-Leiden mutation carriers. Eur. J. Hum. Genet. 2018, 26, 1227–1229. [Google Scholar] [CrossRef] [PubMed]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; DiMaio, D. Human papillomavirus in cervical and head-and-neck cancer. Nat. Clin. Pract. Oncol. 2008, 5, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Luquin, I.; Guarch, R.; Ojer, A.; Pérez-Janices, N.; Martín-Sánchez, E.; Maria-Ruiz, S.; Monreal-Santesteban, I.; Blanco-Fernandez, L.; Pernaut-Leza, E.; Escors, D.; et al. Differential role of gene hypermethylation in adenocarcinomas, squamous cell carcinomas and cervical intraepithelial lesions of the uterine cervix. Pathol. Int. 2015, 65, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Carestiato, F.N.; Afonso, L.A.; Moysés, N.; Almeida Filho, G.L.; Velarde, L.G.; Cavalcanti, S.M. An upward trend in DNA p16ink4a methylation pattern and high risk HPV infection according to the severity of the cervical lesion. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 329–334. [Google Scholar] [CrossRef][Green Version]
- Yang, H.-J.; Liu, V.W.S.; Wang, Y.; Tsang, P.C.K.; Ngan, H.Y.S. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer 2006, 6, 212. [Google Scholar] [CrossRef]
- Guerrero, D.; Guarch, R.; Ojer, A.; Casas, J.M.; Ropero, S.; Mancha, A.; Pesce, C.; Lloveras, B.; Garcia-Bragado, F.; Puras, A. Hypermethylation of the thrombospondin-1 gene is associated with poor prognosis in penile squamous cell carcinoma. BJU Int. 2008, 102, 747–755. [Google Scholar] [CrossRef]
- Ferreux, E.; Lont, A.P.; Horenblas, S.; Gallee, M.P.; Raaphorst, F.M.; von Knebel Doeberitz, M.; Meijer, C.J.; Snijders, P.J. Evidence for at least three alternative mechanisms targeting the p16INK4A/cyclin D/Rb pathway in penile carcinoma, one of which is mediated by high-risk human papillomavirus. J. Pathol. 2003, 201, 109–118. [Google Scholar] [CrossRef]
- Dammann, R.; Li, C.; Yoon, J.H.; Chin, P.L.; Bates, S.; Pfeifer, G.P. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000, 25, 315–319. [Google Scholar] [CrossRef]
- Donninger, H.; Vos, M.D.; Clark, G.J. The RASSF1A tumor suppressor. J. Cell Sci. 2007, 120 Pt 18, 3163–3172. [Google Scholar] [CrossRef]
- Senchenko, V.N.; Kisseljova, N.P.; Ivanova, T.A.; Dmitriev, A.A.; Krasnov, G.S.; Kudryavtseva, A.V.; Panasenko, G.V.; Tsitrin, E.B.; Lerman, M.I.; Kisseljov, F.L.; et al. Novel tumor suppressor candidates on chromosome 3 revealed by NotI-microarrays in cervical cancer. Epigenetics 2013, 8, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Kausar Neyaz, M.; Suresh Kumar, R.; Hussain, S.; Naqvi, S.; Kohaar, I.; Thakur, N.; Kashyap, V.; Das, B.; Akhtar Husain, S.; Bharadwaj, M. Effect of aberrant promoter methylation of FHIT and RASSF1A genes on susceptibility to cervical cancer in a North Indian population. Biomarkers 2008, 13, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Agrawal, S.; Diederichs, S.; Baumer, N.; Becker, A.; Cauvet, T.; Kowski, S.; Beger, C.; Welte, K.; Berdel, W.E.; et al. Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell cycle progression in somatic cells. Oncogene 2005, 24, 2739–2744. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Matzuk, M.M.; Sung, W.K.; Guo, Q.; Wang, P.; Wolgemuth, D.J. Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 1998, 20, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Muller-Tidow, C.; Ji, P.; Diederichs, S.; Potratz, J.; Baumer, N.; Kohler, G.; Cauvet, T.; Choudary, C.; van der Meer, T.; Chan, W.Y.; et al. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol. Cell. Biol. 2004, 24, 8917–8928. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Eijsink, J.J.; Lendvai, A.; Volders, H.H.; Klip, H.; Buikema, H.J.; van Hemel, B.M.; Schuuring, E.; van der Zee, A.G.; Wisman, G.B. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol. Biomarkers Prev. 2009, 18, 3000–3007. [Google Scholar] [CrossRef]
- Kitkumthorn, N.; Yanatatsanajit, P.; Kiatpongsan, S.; Phokaew, C.; Triratanachat, S.; Trivijitsilp, P.; Termrungruanglert, W.; Tresukosol, D.; Niruthisard, S.; Mutirangura, A. Cyclin A1 promoter hypermethylation in human papillomavirus-associated cervical cancer. BMC Cancer 2006, 6, 55. [Google Scholar] [CrossRef]
- Yanatatsaneejit, P.; Chalermchai, T.; Kerekhanjanarong, V.; Shotelersuk, K.; Supiyaphun, P.; Mutirangura, A.; Sriuranpong, V. Promoter hypermethylation of CCNA1, RARRES1, and HRASLS3 in nasopharyngeal carcinoma. Oral Oncol. 2008, 44, 400–406. [Google Scholar] [CrossRef]
- Chujan, S.; Kitkumthorn, N.; Siriangkul, S.; Mutirangura, A. CCNA1 promoter methylation: a potential marker for grading Papanicolaou smear cervical squamous intraepithelial lesions. Asian Pac J Cancer Prev 2014, 15, 7971–7975. [Google Scholar] [CrossRef]
- Hulpiau, P.; van Roy, F. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 2009, 41, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Berx, G.; van Roy, F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a003129. [Google Scholar] [CrossRef] [PubMed]
- Shen-Gunther, J.; Wang, C.-M.; Poage, G.M.; Lin, C.-L.; Perez, L.; Banks, N.A.; Huang, T.H.-M. Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology. Clin. Epigenet. 2016, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Narayan, G.; Arias-Pulido, H.; Koul, S.; Vargas, H.; Zhang, F.F.; Villella, J.; Schneider, A.; Terry, M.B.; Mansukhani, M.; Murty, V.V. Frequent Promoter Methylation of CDH1, DAPK, RARB, and HIC1 Genes in Carcinoma of Cervix Uteri: Its Relationship to Clinical Outcome. Mol. Cancer 2003, 2, 24. [Google Scholar] [CrossRef]
- Flatley, J.E.; McNeir, K.; Balasubramani, L.; Tidy, J.; Stuart, E.L.; Young, T.A.; Powers, H.J. Folate Status and Aberrant DNA Methylation Are Associated with HPV Infection and Cervical Pathogenesis. Cancer Epidemiol. Biomarkers Prev. 2009, 1055–9965. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.-H.; Lin, C.-J.; Liu, C.-J.; Liu, D.-W.; Huang, R.-L.; Ding, D.-C.; Weng, C.-F.; Chu, T.-Y. Global methylation silencing of clustered proto-cadherin genes in cervical cancer: Serving as diagnostic markers comparable to HPV. Cancer Med. 2015, 4, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, A.; Ivarsson, L.; Blassnig, A.; Müller, H.M.; Fiegl, H.; Wiedemair, A.; Müller-Holzner, E.; Goebel, G.; Marth, C.; Widschwendter, M. CDH1 and CDH13 methylation in serum is an independent prognostic marker in cervical cancer patients. Int. J. Cancer 2004, 109, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Narayan, G.; Scotto, L.; Neelakantan, V.; Kottoor, S.H.; Wong, A.H.Y.; Loke, S.-L.; Mansukhani, M.; Pothuri, B.; Wright, J.D.; Kaufmann, A.M.; et al. Protocadherin PCDH10, involved in tumor progression, is a frequent and early target of promoter hypermethylation in cervical cancer. Genes Chromosomes Cancer 2009, 48, 983–992. [Google Scholar] [CrossRef]
- Bhat, S.; Kabekkodu, S.P.; Varghese, V.K.; Chakrabarty, S.; Mallya, S.P.; Rotti, H.; Pandey, D.; Kushtagi, P.; Satyamoorthy, K. Aberrant gene-specific DNA methylation signature analysis in cervical cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Hansel, A.; Steinbach, D.; Greinke, C.; Schmitz, M.; Eiselt, J.; Scheungraber, C.; Gajda, M.; Hoyer, H.; Runnebaum, I.B.; Dürst, M. A promising DNA methylation signature for the triage of high-risk human papillomavirus DNA-positive women. PLoS ONE 2014, 9, e91905. [Google Scholar] [CrossRef]
- Gerecke, C.; Scholtka, B.; Löwenstein, Y.; Fait, I.; Gottschalk, U.; Rogoll, D.; Melcher, R.; Kleuser, B. Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: Putative risk markers for colitis-associated cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jung, E.J.; Lee, H.S.; Kim, M.A.; Kim, W.H. Comparative analysis of DNA methylation between primary and metastatic gastric carcinoma. Oncol. Rep. 2009, 21, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Pulkka, O.P.; Mpindi, J.P.; Tynninen, O.; Nilsson, B.; Kallioniemi, O.; Sihto, H.; Joensuu, H. Clinical relevance of integrin alpha 4 in gastrointestinal stromal tumours. J. Cell. Mol. Med. 2018, 22, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- O’Doherty, C.; Roos, I.M.; Antiguedad, A.; Aransay, A.M.; Hillert, J.; Vandenbroeck, K. ITGA4 polymorphisms and susceptibility to multiple sclerosis. J. Neuroimmunol. 2007, 189, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Kinashi, T. Overview of integrin signaling in the immune system. Methods Mol. Biol. 2012, 757, 261–278. [Google Scholar] [PubMed]
- Rose, D.M.; Han, J.; Ginsberg, M.H. Alpha4 integrins and the immune response. Immunol. Rev. 2002, 186, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.P.; Andersson, E.C.; Scheynius, A.; Marker, O.; Thomsen, A.R. Alpha 4 integrin directs virus-activated CD8+ T cells to sites of infection. J. Immunol. 1995, 154, 5293–5301. [Google Scholar]
- Hilska, M.; Roberts, P.J.; Collan, Y.U.; Laine, V.J.; Kossi, J.; Hirsimaki, P.; Rahkonen, O.; Laato, M. Prognostic significance of matrix metalloproteinases-1, -2, -7 and -13 and tissue inhibitors of metalloproteinases-1, -2, -3 and -4 in colorectal cancer. Int. J. Cancer 2007, 121, 714–723. [Google Scholar] [CrossRef]
- Riddick, A.C.; Shukla, C.J.; Pennington, C.J.; Bass, R.; Nuttall, R.K.; Hogan, A.; Sethia, K.K.; Ellis, V.; Collins, A.T.; Maitland, N.J.; et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br. J. Cancer 2005, 92, 2171–2180. [Google Scholar] [CrossRef]
- Helleman, J.; Jansen, M.P.; Ruigrok-Ritstier, K.; van Staveren, I.L.; Look, M.P.; Meijer-van Gelder, M.E.; Sieuwerts, A.M.; Klijn, J.G.; Sleijfer, S.; Foekens, J.A.; et al. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin. Cancer Res. 2008, 14, 5555–5564. [Google Scholar] [CrossRef]
- Das, A.M.; Bolkestein, M.; van der Klok, T.; Oude Ophuis, C.M.; Vermeulen, C.E.; Rens, J.A.; Dinjens, W.N.; Atmodimedjo, P.N.; Verhoef, C.; Koljenovic, S.; et al. Tissue inhibitor of metalloproteinase-3 (TIMP3) expression decreases during melanoma progression and inhibits melanoma cell migration. Eur. J. Cancer 2016, 66, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Jarzynka, M.J.; Hu, B.; Hui, K.M.; Bar-Joseph, I.; Gu, W.; Hirose, T.; Haney, L.B.; Ravichandran, K.S.; Nishikawa, R.; Cheng, S.Y. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007, 67, 7203–7211. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Fu, H.; Yan, J.; Wang, Y.; Guo, H.; Hao, X.; Xu, X.; Jin, T.; Zhang, N. Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat. Commun. 2013, 4, 1706. [Google Scholar] [CrossRef] [PubMed]
- García, D.; Abba, M.; Briceño, I.; Aristizabal, F.A.; Arregui, A. DNA methylation pattern in high-grade cervical intraepithelial neoplasia and cancer revealed by genome-wide methylation analysis of cervical DNA. Integr. Mol. Med. 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Sato, H.; Miyamoto, T.; Yogev, L.; Namiki, M.; Koh, E.; Hayashi, H.; Sasaki, Y.; Ishikawa, M.; Lamb, D.J.; Matsumoto, N.; et al. Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J. Hum. Genet. 2006, 51, 533–540. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 retrotransposition activity in human genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.; Lutz-Prigge, S.; Moran, J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell 2002, 110, 315–325. [Google Scholar] [CrossRef]
- Inoue, K.; Ichiyanagi, K.; Fukuda, K.; Glinka, M.; Sasaki, H. Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLoS Genet. 2017, 13, e1006926. [Google Scholar] [CrossRef]
- Stanley, M.A. Epithelial cell responses to infection with human papillomavirus. Clin. Microbiol. Rev. 2012, 25, 215–222. [Google Scholar] [CrossRef]
- White, E.A. Manipulation of Epithelial Differentiation by HPV Oncoproteins. Viruses 2019, 11, 369. [Google Scholar] [CrossRef]
- Reuschenbach, M.; Huebbers, C.U.; Prigge, E.S.; Bermejo, J.L.; Kalteis, M.S.; Preuss, S.F.; Seuthe, I.M.; Kolligs, J.; Speel, E.J.; Olthof, N.; et al. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer 2015, 121, 1966–1976. [Google Scholar] [CrossRef] [PubMed]
- Rosl, F.; Arab, A.; Klevenz, B.; zur Hausen, H. The effect of DNA methylation on gene regulation of human papillomaviruses. J. Gen. Virol. 1993, 74 Pt 5, 791–801. [Google Scholar] [CrossRef]
- List, H.J.; Patzel, V.; Zeidler, U.; Schopen, A.; Ruhl, G.; Stollwerk, J.; Klock, G. Methylation sensitivity of the enhancer from the human papillomavirus type 16. J. Biol. Chem. 1994, 269, 11902–11911. [Google Scholar] [PubMed]
- Tan, S.H.; Leong, L.E.; Walker, P.A.; Bernard, H.U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 1994, 68, 6411–6420. [Google Scholar] [CrossRef] [PubMed]
- Chaiwongkot, A.; Vinokurova, S.; Pientong, C.; Ekalaksananan, T.; Kongyingyoes, B.; Kleebkaow, P.; Chumworathayi, B.; Patarapadungkit, N.; Reuschenbach, M.; von Knebel Doeberitz, M. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer 2013, 132, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Woodman, C.B.; Collins, S.I.; Young, L.S. The natural history of cervical HPV infection: Unresolved issues. Nat. Rev. Cancer 2007, 7, 11–22. [Google Scholar] [CrossRef]
- Gammoh, N.; Isaacson, E.; Tomaic, V.; Jackson, D.J.; Doorbar, J.; Banks, L. Inhibition of HPV-16 E7 oncogenic activity by HPV-16 E2. Oncogene 2009, 28, 2299–2304. [Google Scholar] [CrossRef]
- Sanchez, I.E.; Dellarole, M.; Gaston, K.; de Prat Gay, G. Comprehensive comparison of the interaction of the E2 master regulator with its cognate target DNA sites in 73 human papillomavirus types by sequence statistics. Nucleic Acids Res. 2008, 36, 756–769. [Google Scholar] [CrossRef][Green Version]
- Steger, G.; Corbach, S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol. 1997, 71, 50–58. [Google Scholar] [CrossRef]
- Clarke, M.A.; Wentzensen, N.; Mirabello, L.; Ghosh, A.; Wacholder, S.; Harari, A.; Lorincz, A.; Schiffman, M.; Burk, R.D. Human Papillomavirus DNA Methylation as a Potential Biomarker for Cervical Cancer. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 2125–2137. [Google Scholar] [CrossRef]
- O’Connor, M.; Tan, S.-H.; Tan, C.; Bernard, H. YY1 Represses Human Papillomavirus Type 16 Transcription by Quenching AP1 Activity. J. Virol. 1996, 70, 6529–6539. [Google Scholar] [CrossRef] [PubMed]
- Amaro-Filho, S.M.; Pereira Chaves, C.B.; Felix, S.P.; Basto, D.L.; de Almeida, L.M.; Moreira, M.A.M. HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer. Papillomavirus Res. 2018, 5, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Demeret, C. The HPV E2-Host Protein-Protein Interactions: A Complex Hijacking of the Cellular Network. Open Virol. J. 2012, 6, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Grossman, S.R.; Laimins, L.A.; Sigler, P.B. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DMA-binding domain bound to its DNA target. Nature 1992, 359, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Thain, A.; Jenkins, O.; Clarke, A.R.; Gaston, K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996, 70, 7233–7235. [Google Scholar] [CrossRef]
- von Knebel Doeberitz, M.; Prigge, E.S. Role of DNA methylation in HPV associated lesions. Papillomavirus Res. 2019, 7, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Deng, Z.; Pan, X.; Uehara, T.; Suzuki, M.; Xie, M. Effects of Methylation Status of CpG Sites within the HPV16 Long Control Region on HPV16-Positive Head and Neck Cancer Cells. PLoS ONE 2015, 10, e0141245. [Google Scholar] [CrossRef]
- Park, I.S.; Chang, X.; Loyo, M.; Wu, G.; Chuang, A.; Kim, M.S.; Chae, Y.K.; Lyford-Pike, S.; Westra, W.H.; Saunders, J.R.; et al. Characterization of the methylation patterns in human papillomavirus type 16 viral DNA in head and neck cancers. Cancer Prev. Res. 2011, 4, 207–217. [Google Scholar] [CrossRef]
- Balderas-Loaeza, A.; Anaya-Saavedra, G.; Ramirez-Amador, V.A.; Guido-Jimenez, M.C.; Kalantari, M.; Calleja-Macias, I.E.; Bernard, H.U.; Garcia-Carranca, A. Human papillomavirus-16 DNA methylation patterns support a causal association of the virus with oral squamous cell carcinomas. Int. J. Cancer 2007, 120, 2165–2169. [Google Scholar] [CrossRef]
- Badal, V.; Chuang, L.S.H.; Tan, E.H.-H.; Badal, S.; Villa, L.L.; Wheeler, C.M.; Li, B.F.L.; Bernard, H.U. CpG Methylation of Human Papillomavirus Type 16 DNA in Cervical Cancer Cell Lines and in Clinical Specimens: Genomic Hypomethylation Correlates with Carcinogenic Progression. J. Virol. 2003, 77, 6227–6234. [Google Scholar] [CrossRef]
- Mazumder Indra, D.; Singh, R.K.; Mitra, S.; Dutta, S.; Chakraborty, C.; Basu, P.S.; Mondal, R.K.; Roychoudhury, S.; Panda, C.K. Genetic and epigenetic changes of HPV16 in cervical cancer differentially regulate E6/E7 expression and associate with disease progression. Gynecol. Oncol. 2011, 123, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Hublarova, P.; Hrstka, R.; Rotterova, P.; Rotter, L.; Coupkova, M.; Badal, V.; Nenutil, R.; Vojtesek, B. Prediction of human papillomavirus 16 e6 gene expression and cervical intraepithelial neoplasia progression by methylation status. Int. J. Gynecol. Cancer 2009, 19, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.A.; Gradissimo, A.; Schiffman, M.; Lam, J.; Sollecito, C.C.; Fetterman, B.; Lorey, T.; Poitras, N.; Raine-Bennett, T.R.; Castle, P.E.; et al. Human Papillomavirus DNA Methylation as a Biomarker for Cervical Precancer: Consistency across 12 Genotypes and Potential Impact on Management of HPV-Positive Women. Clin. Cancer Res. 2018, 24, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, M.; Calleja-Macias, I.E.; Tewari, D.; Hagmar, B.; Lie, K.; Barrera-Saldana, H.A.; Wiley, D.J.; Bernard, H.U. Conserved Methylation Patterns of Human Papillomavirus Type 16 DNA in Asymptomatic Infection and Cervical Neoplasia. J. Virol. 2004, 78, 12762–12772. [Google Scholar] [CrossRef]
- Wilson, G.A.; Lechner, M.; Koferle, A.; Caren, H.; Butcher, L.M.; Feber, A.; Fenton, T.; Jay, A.; Boshoff, C.; Beck, S. Integrated virus-host methylome analysis in head and neck squamous cell carcinoma. Epigenetics 2013, 8, 953–961. [Google Scholar] [CrossRef]
- Stich, M.; Ganss, L.; Puschhof, J.; Prigge, E.-S.; Reuschenbach, M.; Guiterrez, A.; Vinokurova, S.; von Knebel Doeberitz, M. 5-aza-2’-deoxycytidine (DAC) treatment downregulates the HPV E6 and E7 oncogene expression and blocks neoplastic growth of HPV-associated cancer cells. Oncotarget 2016, 8, 52104–52117. [Google Scholar] [CrossRef]
- Biktasova, A.; Hajek, M.; Sewell, A.; Gary, C.; Bellinger, G.; Deshpande, H.A.; Bhatia, A.; Burtness, B.; Judson, B.; Mehra, S.; et al. Demethylation Therapy as a Targeted Treatment for Human Papillomavirus–Associated Head and Neck Cancer. Clin. Cancer Res. 2017, 23, 7276–7287. [Google Scholar] [CrossRef]
- Bais, M.V. Impact of Epigenetic Regulation on Head and Neck Squamous Cell Carcinoma. J. Dent. Res. 2019, 98, 268–276. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).