How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance
Abstract
:1. Introduction
2. HP1 Family Structure and Function
3. HP1 Post-Translational Modifications
3.1. Phosphorylation
3.2. Acetylation
3.3. Methylation
3.4. Formylation
3.5. Citrullination
3.6. SUMOylation
3.7. Ubiquitination
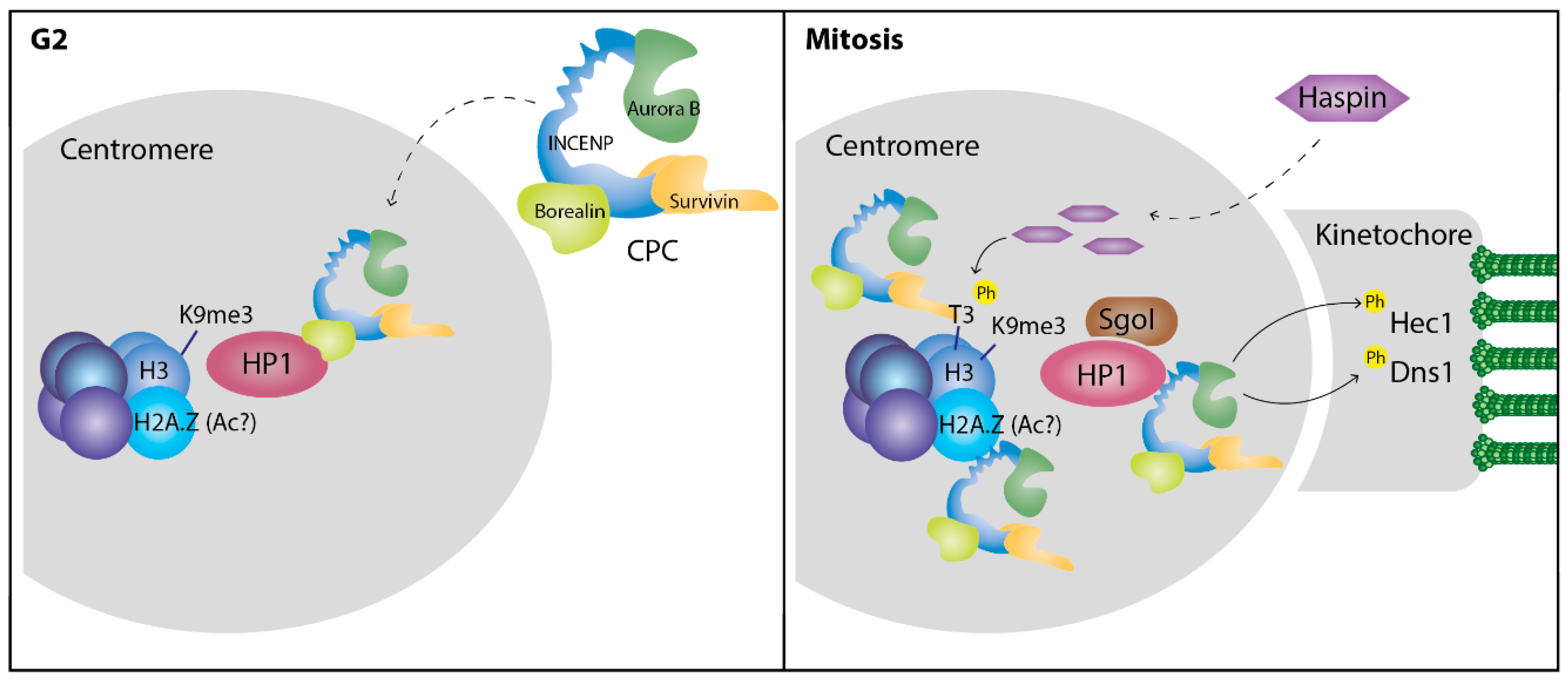
4. HP1 during Mitosis
HP1 Function at the Centromeres
5. Conclusions
Funding
Conflicts of Interest
References
- Armstrong, R.L.; Duronio, R.J. Phasing in heterochromatin during development. Genes Dev. 2019, 33, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, C.L.; Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010, 2, a000596. [Google Scholar] [CrossRef]
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017, 25, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachner, M.; O’carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef]
- Jacobs, S.A.; Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 2002, 295, 2080–2083. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kono, H. Heterochromatin protein 1 (HP1): Interactions with itself and chromatin components. Biophys. Rev. 2020, 12, 387–400. [Google Scholar] [CrossRef]
- Dimitri, P.; Pisano, C. Position effect variegation in Drosophila melanogaster: Relationship between suppression effect and the amount of Y chromosome. Genetics 1989, 122, 793–800. [Google Scholar]
- Mathison, A.; De Assuncao, T.M.; Dsouza, N.R.; Williams, M.; Zimmermann, M.T.; Urrutia, R.; Lomberk, G. Discovery, expression, cellular localization, and molecular properties of a novel, alternative spliced HP1γ isoform, lacking the chromoshadow domain. PLoS ONE 2020, 15, e0217452. [Google Scholar] [CrossRef] [Green Version]
- Minc, E.; Courvalin, J.; Buendia, B. HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Genome Res. 2000, 90, 279–284. [Google Scholar] [CrossRef]
- Horsley, D.; Hutchings, A.; Butcher, G.W.; Singh, P.B. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Genome Res. 1996, 73, 308–311. [Google Scholar] [CrossRef]
- Fanti, L.; Berloco, M.; Piacentini, L.; Pimpinelli, S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica 2003, 117, 135–147. [Google Scholar] [CrossRef]
- Aucott, R.; Bullwinkel, J.; Yu, Y.; Shi, W.; Billur, M.; Brown, J.P.; Menzel, U.; Kioussis, D.; Wang, G.; Reisert, I. HP1-β is required for development of the cerebral neocortex and neuromuscular junctions. J. Cell Biol. 2008, 183, 597–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.P.; Bullwinkel, J.; Baron-Lühr, B.; Billur, M.; Schneider, P.; Winking, H.; Singh, P.B. HP1γ function is required for male germ cell survival and spermatogenesis. Epigenetics Chromatin 2010, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.O.; Cowell, I.G.; Singh, P.B. Mammalian chromodomain proteins: Their role in genome organisation and expression. Bioessays 2000, 22, 124–137. [Google Scholar] [CrossRef]
- Lomberk, G.; Wallrath, L.; Urrutia, R. The heterochromatin protein 1 family. Genome Biol. 2006, 7, 228. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef]
- Murzina, N.; Verreault, A.; Laue, E.; Stillman, B. Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 1999, 4, 529–540. [Google Scholar] [CrossRef]
- Le Douarin, B.; Nielsen, A.L.; Garnier, J.M.; Ichinose, H.; Jeanmougin, F.; Losson, R.; Chambon, P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996, 15, 6701–6715. [Google Scholar] [CrossRef]
- Thiru, A.; Nietlispach, D.; Mott, H.R.; Okuwaki, M.; Lyon, D.; Nielsen, P.R.; Hirshberg, M.; Verreault, A.; Murzina, N.V.; Laue, E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004, 23, 489–499. [Google Scholar] [CrossRef] [Green Version]
- Ainsztein, A.M.; Kandels-Lewis, S.E.; Mackay, A.M.; Earnshaw, W.C. INCENP centromere and spindle targeting: Identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 1998, 143, 1763–1774. [Google Scholar] [CrossRef]
- Liu, X.; Song, Z.; Huo, Y.; Zhang, J.; Zhu, T.; Wang, J.; Zhao, X.; Aikhionbare, F.; Zhang, J.; Duan, H. Chromatin protein HP1α interacts with the mitotic regulator borealin protein and specifies the centromere localization of the chromosomal passenger complex. J. Biol. Chem. 2014, 289, 20638–20649. [Google Scholar] [CrossRef] [Green Version]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Ma, A.; Chow, C.; Horsley, D.; Brown, N.R.; Cowell, I.G.; Singh, P.B. Conservation of heterochromatin protein 1 function. Mol. Cell Biol. 2000, 20, 6970–6983. [Google Scholar] [CrossRef] [Green Version]
- Norwood, L.E.; Grade, S.K.; Cryderman, D.E.; Hines, K.A.; Furiasse, N.; Toro, R.; Li, Y.; Dhasarathy, A.; Kladde, M.P.; Hendrix, M.J. Conserved properties of HP1Hsα. Gene 2004, 336, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Tseng, B.S.; Dormann, H.L.; Ueberheide, B.M.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Funabiki, H.; Allis, C.D. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005, 438, 1116–1122. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Ribeiro, S.; Sennels, L.; Sanchez-Pulido, L.; de Lima Alves, F.; Verheyen, T.; Kelly, D.A.; Ponting, C.P.; Rappsilber, J.; Earnshaw, W.C. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 2011, 21, 328–342. [Google Scholar] [CrossRef] [Green Version]
- de Castro, I.J.; Budzak, J.; Di Giacinto, M.L.; Ligammari, L.; Gokhan, E.; Spanos, C.; Moralli, D.; Richardson, C.; de Las Heras, J.I.; Salatino, S.; et al. Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat. Commun. 2017, 8, 14048. [Google Scholar] [CrossRef]
- Chen, C.C.; Goyal, P.; Karimi, M.M.; Abildgaard, M.H.; Kimura, H.; Lorincz, M.C. H3S10ph broadly marks early-replicating domains in interphase ESCs and shows reciprocal antagonism with H3K9me2. Genome Res. 2018, 28, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Cheung, P.; Allis, C.D.; Sassone-Corsi, P. Signaling to chromatin through histone modifications. Cell 2000, 103, 263–271. [Google Scholar] [CrossRef] [Green Version]
- Mahadevan, L.C.; Willis, A.C.; Barratt, M.J. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 1991, 65, 775–783. [Google Scholar] [CrossRef]
- Wiersma, M.; Bussiere, M.; Halsall, J.A.; Turan, N.; Slany, R.; Turner, B.M.; Nightingale, K.P. Protein kinase Msk1 physically and functionally interacts with the KMT2A/MLL1 methyltransferase complex and contributes to the regulation of multiple target genes. Epigenet. Chromatin 2016, 9, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regnard, C.; Straub, T.; Mitterweger, A.; Dahlsveen, I.K.; Fabian, V.; Becker, P.B. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Li, Y.; Cai, W.; Bao, X.; Girton, J.; Johansen, J.; Johansen, K.M. Histone H3S10 phosphorylation by the JIL-1 kinase in pericentric heterochromatin and on the fourth chromosome creates a composite H3S10phK9me2 epigenetic mark. Chromosoma 2014, 123, 273–280. [Google Scholar] [CrossRef]
- Albig, C.; Wang, C.; Dann, G.P.; Wojcik, F.; Schauer, T.; Krause, S.; Maenner, S.; Cai, W.; Li, Y.; Girton, J. JASPer controls interphase histone H3S10 phosphorylation by chromosomal kinase JIL-1 in Drosophila. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’carroll, D.; Strahl, B.D.; Sun, Z.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Jang, S.M.; Azebi, S.; Soubigou, G.; Muchardt, C. DYRK1A phoshorylates histone H3 to differentially regulate the binding of HP1 isoforms and antagonize HP1-mediated transcriptional repression. EMBO Rep. 2014, 15, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Xhemalce, B.; Kouzarides, T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010, 24, 647–652. [Google Scholar] [CrossRef] [Green Version]
- Clarke, A.S.; Samal, E.; Pillus, L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 2006, 17, 1744–1757. [Google Scholar] [CrossRef] [Green Version]
- Ayrapetov, M.K.; Gursoy-Yuzugullu, O.; Xu, C.; Xu, Y.; Price, B.D. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. USA 2014, 111, 9169–9174. [Google Scholar] [CrossRef] [Green Version]
- Grézy, A.; Chevillard-Briet, M.; Trouche, D.; Escaffit, F. Control of genetic stability by a new heterochromatin compaction pathway involving the Tip60 histone acetyltransferase. Mol. Biol. Cell 2016, 27, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Dong, L.; Tang, M.; Zhang, P.; Zhang, C.; Cao, Z.; Zhu, Q.; Chen, Y.; Wang, H. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018, 46, 7716–7730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiese, M.; Bannister, A.J.; Basu, S.; Boucher, W.; Wohlfahrt, K.; Christophorou, M.A.; Nielsen, M.L.; Klenerman, D.; Laue, E.D.; Kouzarides, T. Citrullination of HP1γ chromodomain affects association with chromatin. Epigenet. Chromatin 2019, 12, 21. [Google Scholar] [CrossRef] [Green Version]
- LeRoy, G.; Weston, J.T.; Zee, B.M.; Young, N.L.; Plazas-Mayorca, M.D.; Garcia, B.A. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol. Cell. Proteom. 2009, 8, 2432–2442. [Google Scholar] [CrossRef] [Green Version]
- Maison, C.; Bailly, D.; Roche, D.; de Oca, R.M.; Probst, A.V.; Vassias, I.; Dingli, F.; Lombard, B.; Loew, D.; Quivy, J. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 2011, 43, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eissenberg, J.C.; Ge, Y.; Hartnett, T. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J. Biol. Chem. 1994, 269, 21315–21321. [Google Scholar] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241. [Google Scholar] [CrossRef]
- Sanulli, S.; Trnka, M.J.; Dharmarajan, V.; Tibble, R.W.; Pascal, B.D.; Burlingame, A.L.; Griffin, P.R.; Gross, J.D.; Narlikar, G.J. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019, 575, 390–394. [Google Scholar] [CrossRef]
- Nishibuchi, G.; Machida, S.; Osakabe, A.; Murakoshi, H.; Hiragami-Hamada, K.; Nakagawa, R.; Fischle, W.; Nishimura, Y.; Kurumizaka, H.; Tagami, H. N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity. Nucleic Acids Res. 2014, 42, 12498–12511. [Google Scholar] [CrossRef] [Green Version]
- Hiragami-Hamada, K.; Shinmyozu, K.; Hamada, D.; Tatsu, Y.; Uegaki, K.; Fujiwara, S.; Nakayama, J. N-terminal phosphorylation of HP1α promotes its chromatin binding. Mol. Cell Biol. 2011, 31, 1186–1200. [Google Scholar] [CrossRef] [Green Version]
- Bryan, L.C.; Weilandt, D.R.; Bachmann, A.L.; Kilic, S.; Lechner, C.C.; Odermatt, P.D.; Fantner, G.E.; Georgeon, S.; Hantschel, O.; Hatzimanikatis, V. Single-molecule kinetic analysis of HP1-chromatin binding reveals a dynamic network of histone modification and DNA interactions. Nucleic Acids Res. 2017, 45, 10504–10517. [Google Scholar] [CrossRef] [PubMed]
- Shimojo, H.; Kawaguchi, A.; Oda, T.; Hashiguchi, N.; Omori, S.; Moritsugu, K.; Kidera, A.; Hiragami-Hamada, K.; Nakayama, J.; Sato, M. Extended string-like binding of the phosphorylated HP1α N-terminal tail to the lysine 9-methylated histone H3 tail. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, T.; Eissenberg, J.C. Phosphorylation of Heterochromatin Protein 1 by Casein Kinase II Is Required for Efficient Heterochromatin Binding inDrosophila. J. Biol. Chem. 1999, 274, 15095–15100. [Google Scholar] [CrossRef] [Green Version]
- Bui, H.T.; Niederwieser, I.; Bird, M.J.; Dai, W.; Brancucci, N.M.; Moes, S.; Jenoe, P.; Lucet, I.S.; Doerig, C.; Voss, T.S. Mapping and functional analysis of heterochromatin protein 1 phosphorylation in the malaria parasite Plasmodium falciparum. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munari, F.; Gajda, M.J.; Hiragami-Hamada, K.; Fischle, W.; Zweckstetter, M. Characterization of the effects of phosphorylation by CK2 on the structure and binding properties of human HP1β. FEBS Lett. 2014, 588, 1094–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, A.; Dohke, K.; Sadaie, M.; Shinmyozu, K.; Nakayama, J.; Urano, T.; Murakami, Y. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009, 23, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, M.; Möröy, T. The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 2005, 37, 726–730. [Google Scholar] [CrossRef]
- Koike, N.; Maita, H.; Taira, T.; Ariga, H.; Iguchi-Ariga, S.M. Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1. FEBS Lett. 2000, 467, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Akaike, Y.; Kuwano, Y.; Nishida, K.; Kurokawa, K.; Kajita, K.; Kano, S.; Masuda, K.; Rokutan, K. Homeodomain-interacting protein kinase 2 regulates DNA damage response through interacting with heterochromatin protein 1γ. Oncogene 2015, 34, 3463–3473. [Google Scholar] [CrossRef]
- Melcher, M.; Schmid, M.; Aagaard, L.; Selenko, P.; Laible, G.; Jenuwein, T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell Biol. 2000, 20, 3728–3741. [Google Scholar] [CrossRef]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Mann, M. N ε-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008, 36, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Uchimura, Y.; Ichimura, T.; Uwada, J.; Tachibana, T.; Sugahara, S.; Nakao, M.; Saitoh, H. Involvement of SUMO modification in MBD1-and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 2006, 281, 23180–23190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maison, C.; Quivy, J.; Almouzni, G. Suv39h1 links the SUMO pathway to constitutive heterochromatin. Mol. Cell. Oncol. 2016, 3, e1225546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maison, C.; Romeo, K.; Bailly, D.; Dubarry, M.; Quivy, J.; Almouzni, G. The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat. Struct. Mol. Biol. 2012, 19, 458. [Google Scholar] [CrossRef]
- Chen, S.; Wang, C.; Sun, L.; Wang, D.; Chen, L.; Huang, Z.; Yang, Q.; Gao, J.; Yang, X.; Chang, J. RAD6 promotes homologous recombination repair by activating the autophagy-mediated degradation of heterochromatin protein HP1. Mol. Cell Biol. 2015, 35, 406–416. [Google Scholar] [CrossRef] [Green Version]
- Hauer, M.H.; Seeber, A.; Singh, V.; Thierry, R.; Sack, R.; Amitai, A.; Kryzhanovska, M.; Eglinger, J.; Holcman, D.; Owen-Hughes, T. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 2017, 24, 99. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, P.; Khanna, R.; Parnaik, V.K. Ubiquitin ligase RNF123 mediates degradation of heterochromatin protein 1α and β in lamin A/C knock-down cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Krishnamoorthy, V.; Khanna, R.; Parnaik, V.K. E3 ubiquitin ligase HECW2 mediates the proteasomal degradation of HP1 isoforms. Biochem. Biophys. Res. Commun. 2018, 503, 2478–2484. [Google Scholar] [CrossRef]
- Grzenda, A.; Leonard, P.; Seo, S.; Mathison, A.J.; Urrutia, G.; Calvo, E.; Iovanna, J.; Urrutia, R.; Lomberk, G. Functional impact of Aurora A-mediated phosphorylation of HP1γ at serine 83 during cell cycle progression. Epigenetics Chromatin 2013, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, K.; Tasaka, H.; Dotsu, M. Molecular behavior in living mitotic cells of human centromere heterochromatin protein HP1α ectopically expressed as a fusion to red fluorescent protein. Cell Struct. Funct. 2001, 26, 705–718. [Google Scholar] [CrossRef] [Green Version]
- Inoue, A.; Hyle, J.; Lechner, M.S.; Lahti, J.M. Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2008, 657, 48–55. [Google Scholar] [CrossRef] [PubMed]
- De Koning, L.; Savignoni, A.; Boumendil, C.; Rehman, H.; Asselain, B.; Sastre-Garau, X.; Almouzni, G. Heterochromatin protein 1α: A hallmark of cell proliferation relevant to clinical oncology. EMBO Mol. Med. 2009, 1, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Sako, K.; Takagaki, K.; Hirayama, Y.; Uchida, K.S.; Herman, J.A.; DeLuca, J.G.; Hirota, T. HP1-assisted Aurora B kinase activity prevents chromosome segregation errors. Dev. Cell 2016, 36, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangasamy, D.; Greaves, I.; Tremethick, D.J. RNA interference demonstrates a novel role for H2A. Z in chromosome segregation. Nat. Struct. Mol. Biol. 2004, 11, 650. [Google Scholar] [CrossRef]
- Fan, J.Y.; Rangasamy, D.; Luger, K.; Tremethick, D.J. H2A. Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 2004, 16, 655–661. [Google Scholar] [CrossRef]
- Greaves, I.K.; Rangasamy, D.; Ridgway, P.; Tremethick, D.J. H2A. Z contributes to the unique 3D structure of the centromere. Proc. Natl. Acad. Sci. USA 2007, 104, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.P.; Tremethick, D.J. The interplay between H2A. Z and H3K9 methylation in regulating HP1α binding to linker histone-containing chromatin. Nucleic Acids Res. 2018, 46, 9353–9366. [Google Scholar] [CrossRef] [Green Version]
- Boyarchuk, E.; Filipescu, D.; Vassias, I.; Cantaloube, S.; Almouzni, G. The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during the cell cycle. J. Cell Sci. 2014, 127, 3347–3359. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, R.S.; Long, H.K.; Swigut, T.; Wysocka, J. Single Amino Acid Change Underlies Distinct Roles of H2A. Z Subtypes in Human Syndrome. Cell 2019, 178, 1421–1436.e24. [Google Scholar] [CrossRef]
- Ruppert, J.G.; Samejima, K.; Platani, M.; Molina, O.; Kimura, H.; Jeyaprakash, A.A.; Ohta, S.; Earnshaw, W.C. HP1α targets the chromosomal passenger complex for activation at heterochromatin before mitotic entry. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Kang, J.; Chaudhary, J.; Dong, H.; Kim, S.; Brautigam, C.A.; Yu, H. Mitotic centromeric targeting of HP1 and its binding to Sgo1 are dispensable for sister-chromatid cohesion in human cells. Mol. Biol. Cell 2011, 22, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Chen, Q.; Yan, H.; Zhang, M.; Liang, C.; Xiang, X.; Pan, X.; Wang, F. Aurora B kinase activity–dependent and–independent functions of the chromosomal passenger complex in regulating sister chromatid cohesion. J. Biol. Chem. 2019, 294, 2021–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, Q.; Chen, Q.; Liang, C.; Yan, H.; Zhang, Z.; Xiang, X.; Zhang, M.; Qi, F.; Zhou, L.; Wang, F. HP1 links centromeric heterochromatin to centromere cohesion in mammals. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Dai, J.; Sullivan, B.A.; Higgins, J.M. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell 2006, 11, 741–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeyaprakash, A.A.; Klein, U.R.; Lindner, D.; Ebert, J.; Nigg, E.A.; Conti, E. Structure of a Survivin–Borealin–INCENP core complex reveals how chromosomal passengers travel together. Cell 2007, 131, 271–285. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Dai, J.; Daum, J.R.; Niedzialkowska, E.; Banerjee, B.; Stukenberg, P.T.; Gorbsky, G.J.; Higgins, J.M. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 2010, 330, 231–235. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, Y.; Honda, T.; Tanno, Y.; Watanabe, Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 2010, 330, 239–243. [Google Scholar] [CrossRef]
- Yamagishi, Y.; Sakuno, T.; Shimura, M.; Watanabe, Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 2008, 455, 251–255. [Google Scholar] [CrossRef]
- Nonaka, N.; Kitajima, T.; Yokobayashi, S.; Xiao, G.; Yamamoto, M.; Grewal, S.I.; Watanabe, Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.; Maure, J.; Partridge, J.F.; Genier, S.; Javerzat, J.; Allshire, R.C. Requirement of heterochromatin for cohesion at centromeres. Science 2001, 294, 2539–2542. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.; Rodríguez-Corsino, M.; Losada, A. Heterochromatin protein 1 (HP1) proteins do not drive pericentromeric cohesin enrichment in human cells. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kniola, B.; O’Toole, E.; McIntosh, J.R.; Mellone, B.; Allshire, R.; Mengarelli, S.; Hultenby, K.; Ekwall, K. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 2001, 12, 2767–2775. [Google Scholar] [CrossRef]
- Hayakawa, T.; Haraguchi, T.; Masumoto, H.; Hiraoka, Y. Cell cycle behavior of human HP1 subtypes: Distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003, 116, 3327–3338. [Google Scholar] [CrossRef] [Green Version]
- Nishibuchi, G.; Machida, S.; Nakagawa, R.; Yoshimura, Y.; Hiragami-Hamada, K.; Abe, Y.; Kurumizaka, H.; Tagami, H.; Nakayama, J. Mitotic phosphorylation of HP1α regulates its cell cycle-dependent chromatin binding. J. Biochem. 2019, 165, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.M.; Mathison, A.J.; Christensen, T.; Greipp, P.T.; Knutson, D.L.; Klee, E.W.; Zimmermann, M.T.; Iovanna, J.; Lomberk, G.A.; Urrutia, R.A. Aurora kinase B-phosphorylated HP1α functions in chromosomal instability. Cell Cycle 2019, 18, 1407–1421. [Google Scholar] [CrossRef]
- Chakraborty, A.; Prasanth, K.V.; Prasanth, S.G. Dynamic phosphorylation of HP1α regulates mitotic progression in human cells. Nat. Commun. 2014, 5, 3445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, A.; Prasanth, S.G. Phosphorylation–dephosphorylation cycle of HP1α governs accurate mitotic progression. Cell Cycle 2014, 13, 1663–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sales-Gil, R.; Vagnarelli, P. How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance. Cells 2020, 9, 1460. https://doi.org/10.3390/cells9061460
Sales-Gil R, Vagnarelli P. How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance. Cells. 2020; 9(6):1460. https://doi.org/10.3390/cells9061460
Chicago/Turabian StyleSales-Gil, Raquel, and Paola Vagnarelli. 2020. "How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance" Cells 9, no. 6: 1460. https://doi.org/10.3390/cells9061460
APA StyleSales-Gil, R., & Vagnarelli, P. (2020). How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance. Cells, 9(6), 1460. https://doi.org/10.3390/cells9061460




