Inhibition of Transglutaminase 2 but Not of MDM2 Has a Significant Therapeutic Effect on Renal Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Culture
2.3. Western Blot Analysis
2.4. Real-Time Apoptosis Assay
2.5. Preclinical Xenograft Tumor Models
2.6. Automated Immunohistochemistry
2.7. Statistical Analysis
3. Results
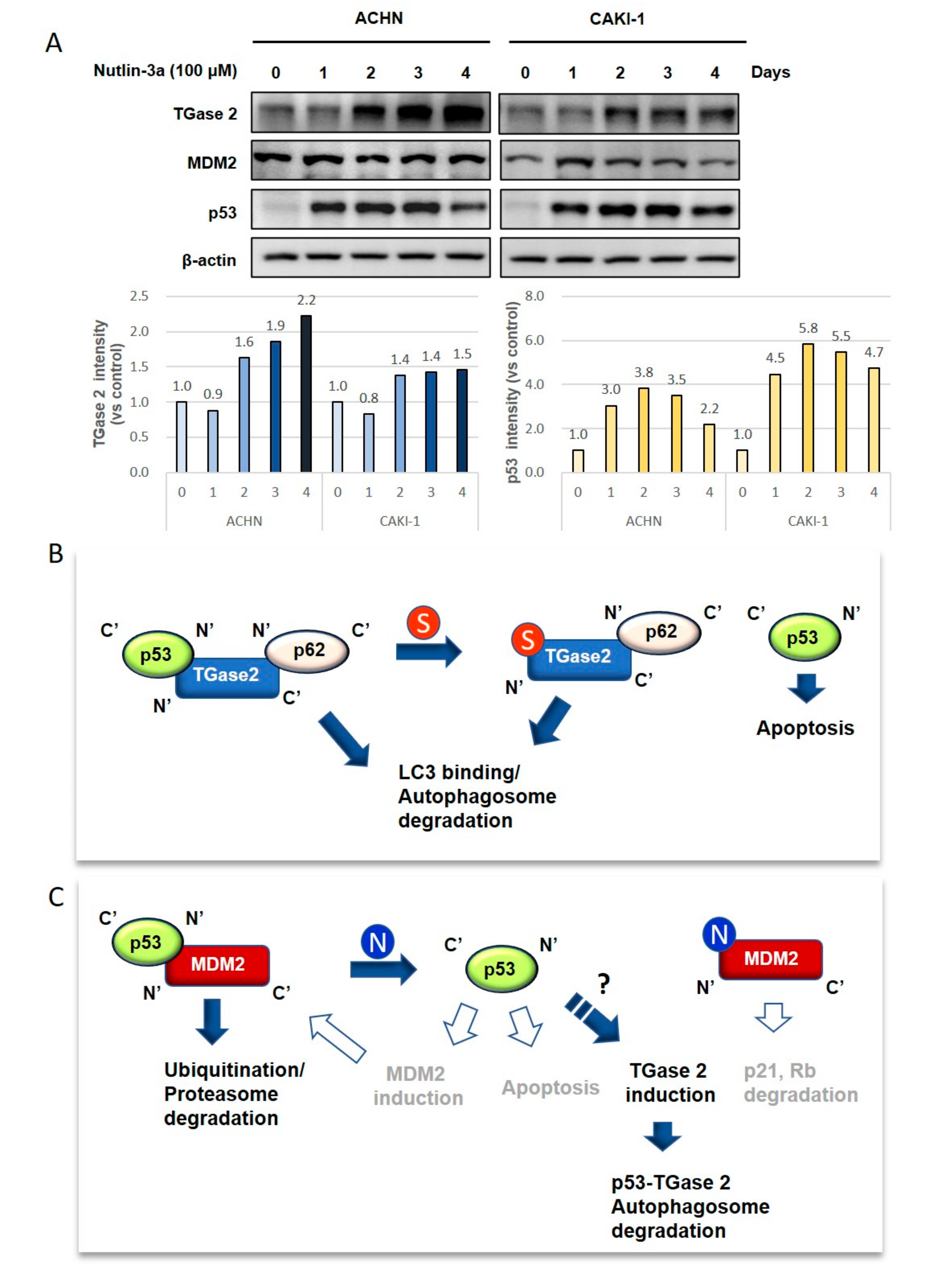
3.1. TGase 2 and MDM2 Suppress p53 in RCC
3.2. p53 in RCC is Stabilized by Treatment with Streptonigrin and Nutlin-3a
3.3. TGase 2 mRNA Levels Increase in RCC
3.4. Inhibiting TGase 2 Suppresses the Growth of RCC in a Xenograft Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Polanski, R.; Noon, A.P.; Blaydes, J.; Phillips, A.; Rubbi, C.P.; Parsons, K.; Vlatkovic, N.; Boyd, M.T. Senescence induction in renal carcinoma cells by Nutlin-3: a potential therapeutic strategy based on MDM2 antagonism. Cancer Lett. 2014, 353, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Lee, J.S.; Hong, D.; Lee, S.H.; Kim, N.; Lee, W.K.; Sung, T.W.; Gong, Y.D.; Kim, S.Y. Renal cell carcinoma escapes death by p53 depletion through transglutaminase 2-chaperoned autophagy. Cell Death Dis. 2016, 7, e2163. [Google Scholar] [CrossRef] [PubMed]
- Oliner, J.D.; Kinzler, K.W.; Meltzer, P.S.; George, D.L.; Vogelstein, B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992, 358, 80–83. [Google Scholar] [CrossRef]
- Iwakuma, T.; Lozano, G. MDM2, an introduction. Mol. Cancer Res. 2003, 1, 993–1000. [Google Scholar]
- Capoulade, C.; Bressac-de Paillerets, B.; Lefrere, I.; Ronsin, M.; Feunteun, J.; Tursz, T.; Wiels, J. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene 1998, 16, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Leng, P.; Brown, D.R.; Shivakumar, C.V.; Deb, S.; Deb, S.P. N-terminal 130 amino acids of MDM2 are sufficient to inhibit p53-mediated transcriptional activation. Oncogene 1995, 10, 1275–1282. [Google Scholar] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.; Aguilar, A.; Bernard, D.; Yang, C.Y. Targeting the MDM2-p53 Protein-Protein Interaction for New Cancer Therapy: Progress and Challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a026245. [Google Scholar] [CrossRef]
- Tonsing-Carter, E.; Bailey, B.J.; Saadatzadeh, M.R.; Ding, J.; Wang, H.; Sinn, A.L.; Peterman, K.M.; Spragins, T.K.; Silver, J.M.; Sprouse, A.A.; et al. Potentiation of Carboplatin-Mediated DNA Damage by the Mdm2 Modulator Nutlin-3a in a Humanized Orthotopic Breast-to-Lung Metastatic Model. Mol. Cancer Ther. 2015, 14, 2850–2863. [Google Scholar] [CrossRef]
- Vu, B.; Wovkulich, P.; Pizzolato, G.; Lovey, A.; Ding, Q.; Jiang, N.; Liu, J.J.; Zhao, C.; Glenn, K.; Wen, Y.; et al. Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development. ACS Med. Chem. Lett. 2013, 4, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Rew, Y.; Sun, D.; Gonzalez-Lopez De Turiso, F.; Bartberger, M.D.; Beck, H.P.; Canon, J.; Chen, A.; Chow, D.; Deignan, J.; Fox, B.M.; et al. Structure-based design of novel inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2012, 55, 4936–4954. [Google Scholar] [CrossRef] [PubMed]
- Holzer, P.; Masuya, K.; Furet, P.; Kallen, J.; Valat-Stachyra, T.; Ferretti, S.; Berghausen, J.; Bouisset-Leonard, M.; Buschmann, N.; Pissot-Soldermann, C.; et al. Discovery of a Dihydroisoquinolinone Derivative (NVP-CGM097): A Highly Potent and Selective MDM2 Inhibitor Undergoing Phase 1 Clinical Trials in p53wt Tumors. J. Med. Chem. 2015, 58, 6348–6358. [Google Scholar] [CrossRef]
- Kang, M.H.; Reynolds, C.P.; Kolb, E.A.; Gorlick, R.; Carol, H.; Lock, R.; Keir, S.T.; Maris, J.M.; Wu, J.; Lyalin, D.; et al. Initial Testing (Stage 1) of MK-8242-A Novel MDM2 Inhibitor-by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer 2016, 63, 1744–1752. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, W.K.; Kim, N.; Kang, J.H.; Kim, K.H.; Kim, S.G.; Lee, J.S.; Lee, S.; Lee, J.; Joo, J.; et al. Renal Cell Carcinoma Is Abrogated by p53 Stabilization through Transglutaminase 2 Inhibition. Cancers 2018, 10, 455. [Google Scholar] [CrossRef] [PubMed]
- Ku, B.M.; Kim, D.S.; Kim, K.H.; Yoo, B.C.; Kim, S.H.; Gong, Y.D.; Kim, S.Y. Transglutaminase 2 inhibition found to induce p53 mediated apoptosis in renal cell carcinoma. FASEB J. 2013, 27, 3487–3495. [Google Scholar] [CrossRef]
- Ku, B.M.; Kim, S.J.; Kim, N.; Hong, D.; Choi, Y.B.; Lee, S.H.; Gong, Y.D.; Kim, S.Y. Transglutaminase 2 inhibitor abrogates renal cell carcinoma in xenograft models. J. Cancer Res. Clin. Oncol. 2014, 140, 757–767. [Google Scholar] [CrossRef]
- Kang, J.H.; Lee, S.H.; Cheong, H.; Lee, C.H.; Kim, S.Y. Transglutaminase 2 Promotes Autophagy by LC3 Induction through p53 Depletion in Cancer Cell. Biomol. Ther. 2019, 27, 34–40. [Google Scholar] [CrossRef]
- Folk, J.E. Mechanism and basis for specificity of transglutaminase-catalyzed epsilon-(gamma-glutamyl) lysine bond formation. Adv. Enzymol. Relat. Areas Mol. Biol. 1983, 54, 1–56. [Google Scholar]
- Keillor, J.W.; Clouthier, C.M.; Apperley, K.Y.P.; Akbar, A.; Mulani, A. Acyl transfer mechanisms of tissue transglutaminase. Bioorg. Chem. 2014, 57, 186–197. [Google Scholar] [CrossRef]
- Folk, J.E. Mechanism of action of guinea pig liver transglutaminase. VI. Order of substrate addition. J. Biol. Chem. 1969, 244, 3707–3713. [Google Scholar] [PubMed]
- Folk, J.E. Structure and catalytic properties of hepatic transglutaminase. Ann. N. Y. Acad. Sci. 1972, 202, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Park, S.S.; Nam, B.H.; Kim, I.H.; Kim, S.Y. Reversal of drug resistance in breast cancer cells by transglutaminase 2 inhibition and nuclear factor-kappaB inactivation. Cancer Res. 2006, 66, 10936–10943. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Choi, Y.B.; Han, B.G.; Park, S.Y.; Jeon, Y.; Kim, D.H.; Ahn, E.R.; Shin, J.E.; Lee, B.I.; Lee, H.; et al. Cancer cells promote survival through depletion of the von Hippel-Lindau tumor suppressor by protein crosslinking. Oncogene 2011, 30, 4780–4790. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Lee, S.H.; Kim, S.Y. Discovery of a novel target for renal cell carcinoma: transglutaminase 2. Cell Death Dis. 2016, 7, e2200. [Google Scholar] [CrossRef]
- Warburton, H.E.; Brady, M.; Vlatkovic, N.; Linehan, W.M.; Parsons, K.; Boyd, M.T. p53 regulation and function in renal cell carcinoma. Cancer Res. 2005, 65, 6498–6503. [Google Scholar] [CrossRef]
- Begus-Nahrmann, Y.; Lechel, A.; Obenauf, A.C.; Nalapareddy, K.; Peit, E.; Hoffmann, E.; Schlaudraff, F.; Liss, B.; Schirmacher, P.; Kestler, H.; et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat. Genet. 2009, 41, 1138–1143. [Google Scholar] [CrossRef]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993, 7, 1126–1132. [Google Scholar] [CrossRef]
- Nag, S.; Zhang, X.; Srivenugopal, K.S.; Wang, M.H.; Wang, W.; Zhang, R. Targeting MDM2-p53 interaction for cancer therapy: are we there yet? Curr. Med. Chem. 2014, 21, 553–574. [Google Scholar] [CrossRef]
- Li, Q.; Lozano, G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 2013, 19, 34–41. [Google Scholar] [CrossRef]
- Bouska, A.; Eischen, C.M. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem. Sci. 2009, 34, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Dean, D.C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000, 14, 2393–2409. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Keillor, J.W. A Precision Strategy to Cure Renal Cell Carcinoma by Targeting Transglutaminase 2. Int. J. Mol. Sci. 2020, 21, 2493. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Harrison, C. Angiogenesis: A deeper understanding of VEGFR inhibitors. Nat. Rev. Cancer 2012, 12, 735. [Google Scholar] [CrossRef]
- Papaetis, G.S.; Syrigos, K.N. Sunitinib: a multitargeted receptor tyrosine kinase inhibitor in the era of molecular cancer therapies. BioDrugs 2009, 23, 377–389. [Google Scholar] [CrossRef]
- Sui, X.; Chen, R.; Wang, Z.; Huang, Z.; Kong, N.; Zhang, M.; Han, W.; Lou, F.; Yang, J.; Zhang, Q.; et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013, 4, e838. [Google Scholar] [CrossRef]
- Hagiwara, N.; Watanabe, M.; Iizuka-Ohashi, M.; Yokota, I.; Toriyama, S.; Sukeno, M.; Tomosugi, M.; Sowa, Y.; Hongo, F.; Mikami, K.; et al. Mevalonate pathway blockage enhances the efficacy of mTOR inhibitors with the activation of retinoblastoma protein in renal cell carcinoma. Cancer Lett. 2018, 431, 182–189. [Google Scholar] [CrossRef]
- Cao, Q.; Bai, P. Role of Autophagy in Renal Cancer. J. Cancer 2019, 10, 2501–2509. [Google Scholar] [CrossRef]
- Grimaldi, A.; Santini, D.; Zappavigna, S.; Lombardi, A.; Misso, G.; Boccellino, M.; Desiderio, V.; Vitiello, P.P.; Di Lorenzo, G.; Zoccoli, A.; et al. Antagonistic effects of chloroquine on autophagy occurrence potentiate the anticancer effects of everolimus on renal cancer cells. Cancer Biol. Ther. 2015, 16, 567–579. [Google Scholar] [CrossRef]
- Cao, J.; Huang, W. Compensatory Increase of Transglutaminase 2 Is Responsible for Resistance to mTOR Inhibitor Treatment. PLoS One 2016, 11, e0149388. [Google Scholar] [CrossRef] [PubMed]
- D’Eletto, M.; Farrace, M.G.; Falasca, L.; Reali, V.; Oliverio, S.; Melino, G.; Griffin, M.; Fimia, G.M.; Piacentini, M. Transglutaminase 2 is involved in autophagosome maturation. Autophagy 2009, 5, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, N.; Kim, S.J.; Song, J.; Gong, Y.D.; Kim, S.Y. Anti-cancer effect of a quinoxaline derivative GK13 as a transglutaminase 2 inhibitor. J. Cancer Res. Clin. Oncol. 2013, 139, 1279–1294. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y. New Insights into Development of Transglutaminase 2 Inhibitors as Pharmaceutical Lead Compounds. Med. Sci. 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kang, J.H.; Lee, W.K.; Kim, S.G.; Lee, J.S.; Lee, S.H.; Park, J.B.; Kim, K.H.; Gong, Y.D.; Hwang, K.Y.; et al. Allosteric inhibition site of transglutaminase 2 is unveiled in the N terminus. Amino Acids 2018, 50, 1583–1594. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.H.; Lee, S.-H.; Lee, J.-S.; Oh, S.-J.; Ha, J.S.; Choi, H.-J.; Kim, S.-Y. Inhibition of Transglutaminase 2 but Not of MDM2 Has a Significant Therapeutic Effect on Renal Cell Carcinoma. Cells 2020, 9, 1475. https://doi.org/10.3390/cells9061475
Kang JH, Lee S-H, Lee J-S, Oh S-J, Ha JS, Choi H-J, Kim S-Y. Inhibition of Transglutaminase 2 but Not of MDM2 Has a Significant Therapeutic Effect on Renal Cell Carcinoma. Cells. 2020; 9(6):1475. https://doi.org/10.3390/cells9061475
Chicago/Turabian StyleKang, Joon Hee, Seon-Hyeong Lee, Jae-Seon Lee, Su-Jin Oh, Ji Sun Ha, Hyun-Jung Choi, and Soo-Youl Kim. 2020. "Inhibition of Transglutaminase 2 but Not of MDM2 Has a Significant Therapeutic Effect on Renal Cell Carcinoma" Cells 9, no. 6: 1475. https://doi.org/10.3390/cells9061475
APA StyleKang, J. H., Lee, S.-H., Lee, J.-S., Oh, S.-J., Ha, J. S., Choi, H.-J., & Kim, S.-Y. (2020). Inhibition of Transglutaminase 2 but Not of MDM2 Has a Significant Therapeutic Effect on Renal Cell Carcinoma. Cells, 9(6), 1475. https://doi.org/10.3390/cells9061475



