Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Access
2.2. Mapping Small RNA Reads
2.3. Analyses of Ping-Pong Signature
2.4. Repeat Masking
2.5. BLASTN Analyses
2.6. SnetDNA Consensus Sequences
2.7. Bioinformatics
2.8. snetDNAs and Genomic Annotations
2.9. Expression of Genes Containing snetDNAs
2.10. Accession Numbers
3. Results
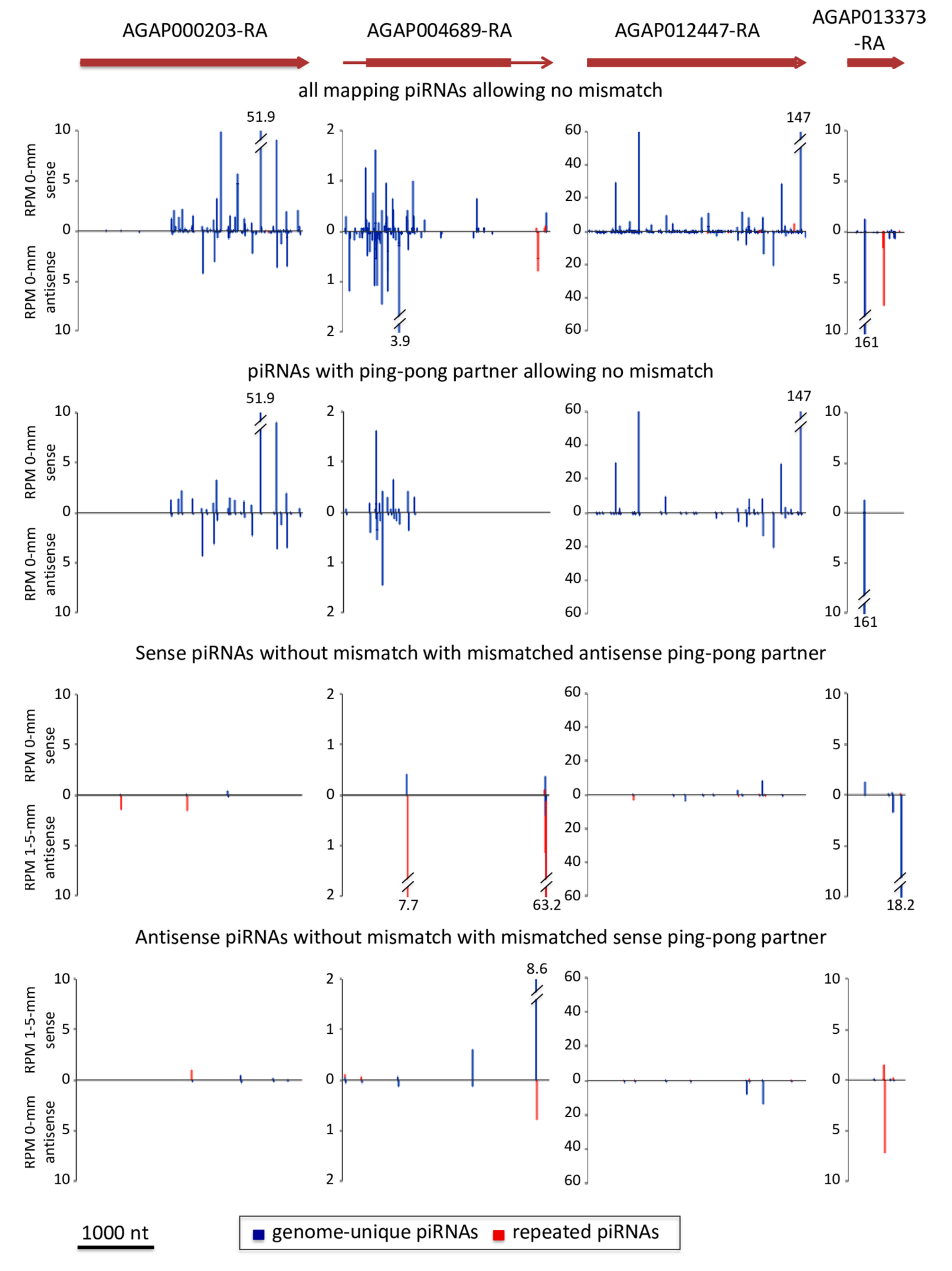
3.1. Genes May Give Rise to piRNAs Implicated in Ping-Pong Amplification in An. gambiae Ovaries
3.2. piRNAs from Genomic Repeats May Fuel mRNAs from Distant Loci into the Ping-Pong Cycle of piRNA Amplification and Create “Ping-Pong Networks”
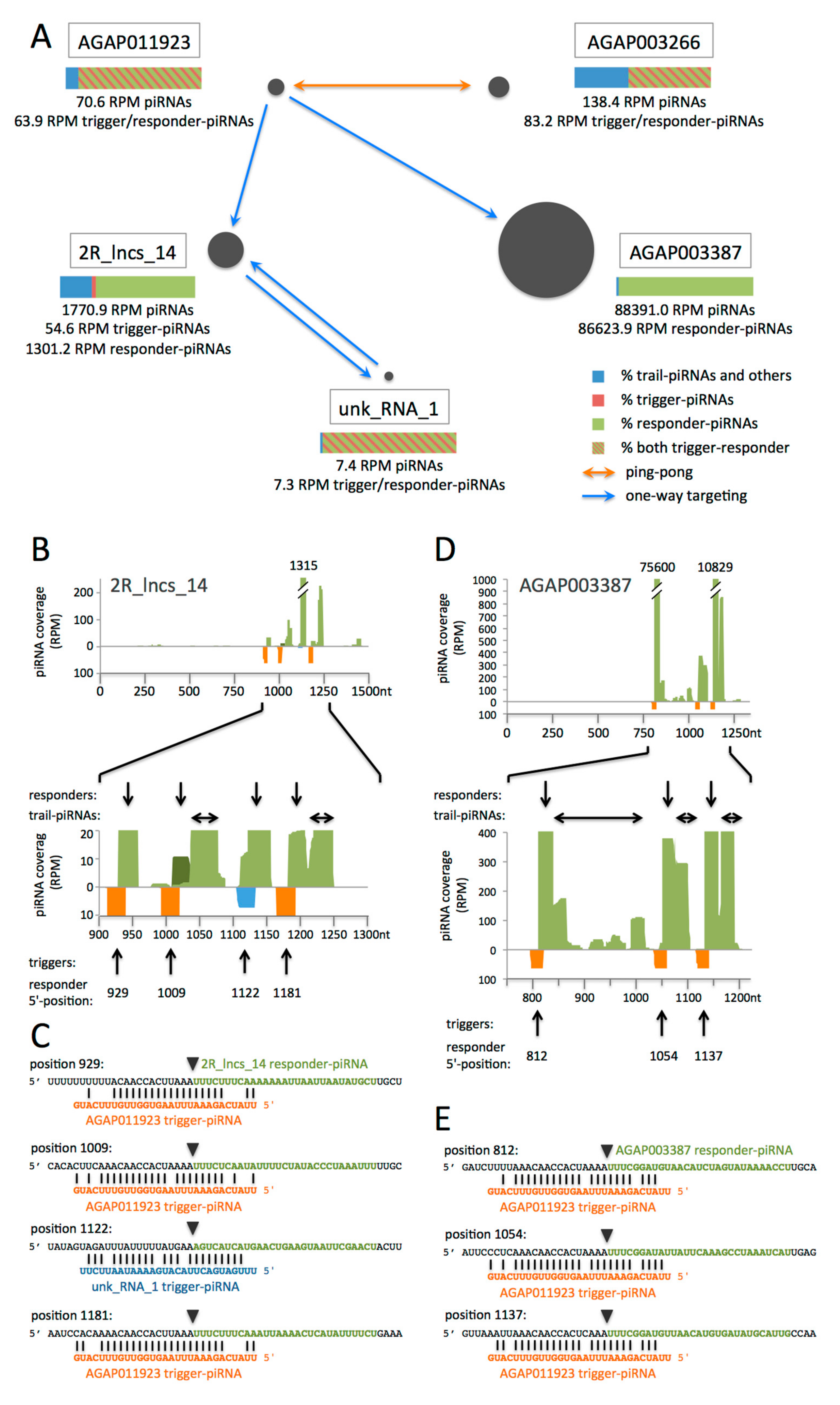
3.3. piRNAs from Genes and lncRNAs May Direct Other Transcripts to Phased piRNA Biogenesis, thus Creating Networks of Inter-Regulation
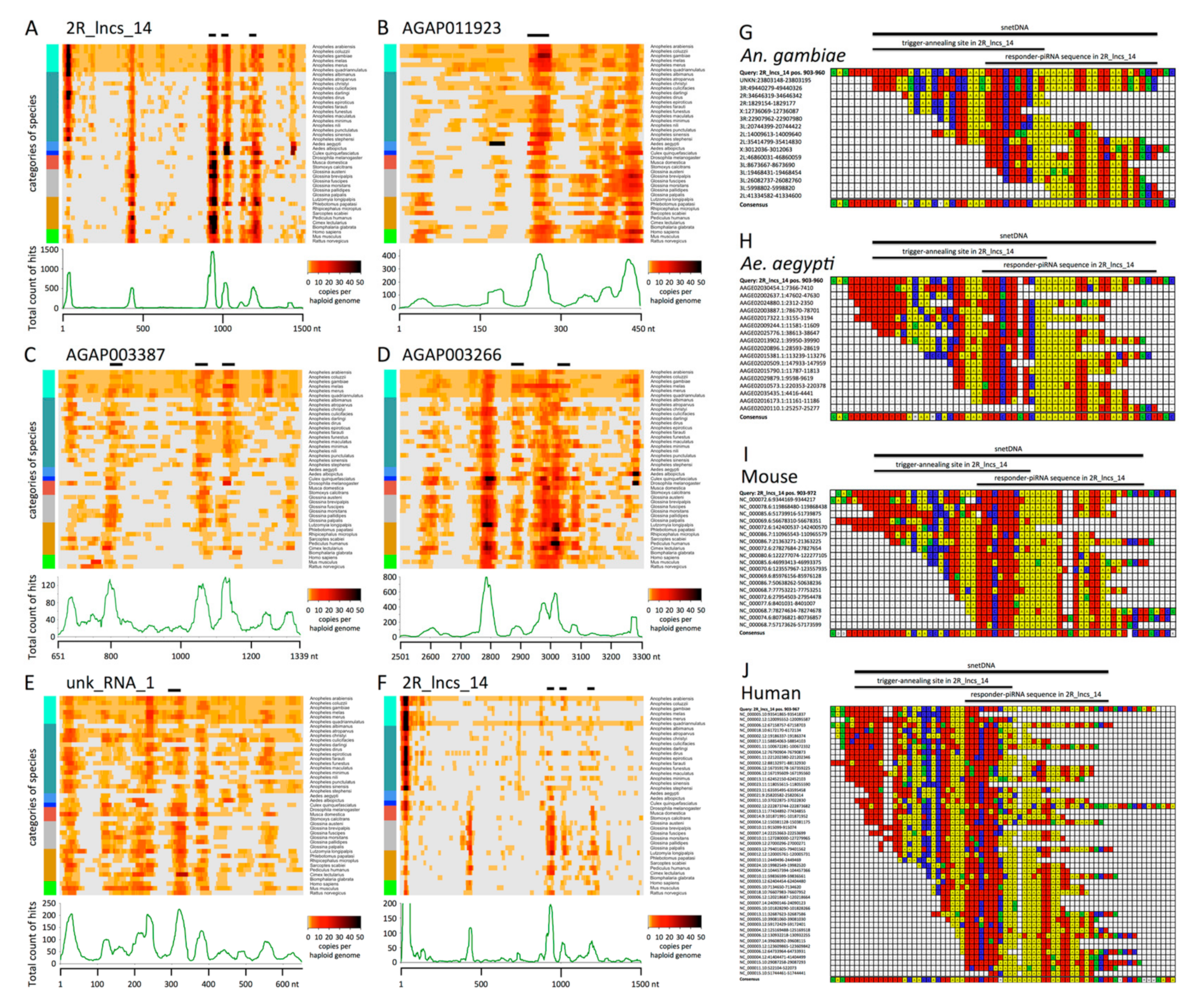
3.4. Genomic Sequences Producing piRNAs Engaged in Networks are Repeated and Conserved from Insects to Mammals
3.5. SnetDNAs from the Aedes aegypti Mosquito Produce piRNAs and Establish Networks
3.6. The Characteristics of snetDNAs in Mouse and Human Suggest that snetDNAs Derive from Ancient Transposable Elements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yoshikawa, M.; Han, B.W.; Izumi, N.; Tomari, Y.; Weng, Z.; Zamore, P.D. The Initial Uridine of Primary piRNAs Does Not Create the Tenth Adenine that Is the Hallmark of Secondary piRNAs. Mol. Cell 2014, 56, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Handler, D.; Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef]
- Gan, H.; Lin, X.; Zhang, Z.; Zhang, W.; Liao, S.; Wang, L.; Han, C. piRNA profiling during specific stages of mouse spermatogenesis. RNA 2011, 17, 1191–1203. [Google Scholar] [CrossRef][Green Version]
- Sarkar, A.; Volff, J.-N.; Vaury, C. piRNAs and their diverse roles: A transposable element-driven tactic for gene regulation? FASEB J. 2017, 31, 436–446. [Google Scholar] [CrossRef]
- Yamtich, J.; Heo, S.-J.; Dhahbi, J.; Martin, D.I.; Boffelli, D. piRNA-like small RNAs mark extended 3′UTRs present in germ and somatic cells. BMC Genom. 2015, 16, 462. [Google Scholar] [CrossRef]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A Broadly Conserved Pathway Generates 3′UTR-Directed Primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef]
- Bouhouche, K.; Gout, J.-F.; Kapusta, A.; Bétermier, M.; Meyer, E. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res. 2011, 39, 4249–4264. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Gu, W.; Shirayama, M.; Youngman, E.; Conte, D.; Mello, C.C. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell 2012, 150, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; Chuva de Sousa Lopes, S.M.; van der Westerlaken, L.A.J.; Zischler, H.; Butter, F.; Roelen, B.A.J.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef] [PubMed]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A Role for Neuronal piRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef]
- Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; Desvignes, J.-P.; et al. Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef]
- Watanabe, T.; Cheng, E.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380. [Google Scholar] [CrossRef]
- Ku, H.-Y.; Lin, H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 2014, 1, 205–218. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar] [CrossRef]
- George, P.; Jensen, S.; Pogorelcnik, R.; Lee, J.; Xing, Y.; Brasset, E.; Vaury, C.; Sharakhov, I.V. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenet. Chromatin 2015, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Castellano, L.; Rizzi, E.; Krell, J.; Di Cristina, M.; Galizi, R.; Mori, A.; Tam, J.; De Bellis, G.; Stebbing, J.; Crisanti, A.; et al. The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs. BMC Genom. 2015, 16, 100. [Google Scholar] [CrossRef] [PubMed]
- Biryukova, I.; Ye, T. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genom. 2015, 16, 278. [Google Scholar] [CrossRef] [PubMed]
- Arensburger, P.; Hice, R.H.; Wright, J.A.; Craig, N.L.; Atkinson, P.W. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genom. 2011, 12, 606. [Google Scholar] [CrossRef]
- Ichiyanagi, T.; Ichiyanagi, K.; Ogawa, A.; Kuramochi-Miyagawa, S.; Nakano, T.; Chuma, S.; Sasaki, H.; Udono, H. HSP90α plays an important role in piRNA biogenesis and retrotransposon repression in mouse. Nucleic Acids Res. 2014, 42, 11903–11911. [Google Scholar] [CrossRef]
- Reuter, M.; Berninger, P.; Chuma, S.; Shah, H.; Hosokawa, M.; Funaya, C.; Antony, C.; Sachidanandam, R.; Pillai, R.S. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 2011, 480, 264–267. [Google Scholar] [CrossRef]
- Pogorelcnik, R.; Vaury, C.; Pouchin, P.; Jensen, S.; Brasset, E. sRNAPipe: A Galaxy-based pipeline for bioinformatic in-depth exploration of small RNAseq data. Mob. DNA 2018, 9, 25. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Dufourt, J.; Pouchin, P.; Peyret, P.; Brasset, E.; Vaury, C. NucBase, an easy to use read mapper for small RNAs. Mob. DNA 2013, 4, 1. [Google Scholar] [CrossRef]
- Giraldo-Calderon, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; The VectorBase Consortium; Madey, G.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, A.M.; Waterhouse, R.M.; Muskavitch, M.A. Long non-coding RNA discovery across the genus anopheles reveals conserved secondary structures within and beyond the Gambiae complex. BMC Genom. 2015, 16, 337. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Grentzinger, T.; Armenise, C.; Brun, C.; Mugat, B.; Serrano, V.; Pelisson, A.; Chambeyron, S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012, 22, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Favorov, A.; Mularoni, L.; Cope, L.M.; Medvedeva, Y.; Mironov, A.A.; Makeev, V.J.; Wheelan, S.J. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 2012, 8, e1002529. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Tuteja, G. TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics 2019, 35, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Shen, Y.; Yue, F.; McCleary, D.F.; Ye, Z.; Edsall, L.; Kuan, S.; Wagner, U.; Dixon, J.; Lee, L.; Lobanenkov, V.V.; et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012, 488, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Gomez-Diaz, E.; Rivero, A.; Chandre, F.; Corces, V.G. Insights into the epigenomic landscape of the human malaria vector Anopheles gambiae. Front. Genet. 2014, 5, 277. [Google Scholar]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Arensburger, P.; Hice, R.H.; Zhou, L.; Smith, R.C.; Tom, A.C.; Wright, J.A.; Knapp, J.; O’Brochta, D.A.; Craig, N.L.; Atkinson, P.W. Phylogenetic and Functional Characterization of the hAT Transposon Superfamily. Genetics 2011, 188, 45–57. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of piRNAs in the central nervous system. RNA 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.-Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.-H.; Dai, S.-Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Kang, J.-Y.; Gou, L.-T.; Wang, J.; Xue, Y.; Skogerboe, G.; Dai, P.; Huang, D.-W.; Chen, R.; Fu, X.-D.; et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015, 25, 193–207. [Google Scholar] [CrossRef]
- Campbell, C.L.; Black, W.C.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom. 2008, 9, 425. [Google Scholar] [CrossRef]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jensen, S.; Brasset, E.; Parey, E.; Roest Crollius, H.; Sharakhov, I.V.; Vaury, C. Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs. Cells 2020, 9, 1491. https://doi.org/10.3390/cells9061491
Jensen S, Brasset E, Parey E, Roest Crollius H, Sharakhov IV, Vaury C. Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs. Cells. 2020; 9(6):1491. https://doi.org/10.3390/cells9061491
Chicago/Turabian StyleJensen, Silke, Emilie Brasset, Elise Parey, Hugues Roest Crollius, Igor V. Sharakhov, and Chantal Vaury. 2020. "Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs" Cells 9, no. 6: 1491. https://doi.org/10.3390/cells9061491
APA StyleJensen, S., Brasset, E., Parey, E., Roest Crollius, H., Sharakhov, I. V., & Vaury, C. (2020). Conserved Small Nucleotidic Elements at the Origin of Concerted piRNA Biogenesis from Genes and lncRNAs. Cells, 9(6), 1491. https://doi.org/10.3390/cells9061491






