Isoleucine 44 Hydrophobic Patch Controls Toxicity of Unanchored, Linear Ubiquitin Chains through NF-κB Signaling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Fly Generation
2.3. Drosophila Stocks and Procedures
2.4. Western Blotting
2.5. Immunoprecipitation and In Vitro Deubiquitination
2.6. Soluble/Insoluble Fractionation
2.7. Quantitative Real-Time PCR
- rp49 (F: AGATCGTGAAGAAGCGCACCAAG; R: CACCAGGAACTTCTTGAATCCGG),
- Relish (F: ATGAACTTGAACCAGGTGCGG; R: TGCCGACTTGCGGTTATTGA),
- Dif (F: CAGAGTTCCAACCCACGGAC; R: AGGAGTTCTGGATTCGGGTAGT),
- dorsal (F: AGAGCCCGCCAAGGTTTTT; R: TGCCATCCTGGTGGTCATTC),
- cactus (F: ATCCAACGACAAAGCGGTCA; R: GATTTTCCCTCCCTGGCGTTA),
- and kenny (F: TACCTCGCGCTAAAGAGCAC; R: CAGCTCTTGGTTTTCCACGC).
2.8. Subcellular Fractionation
2.9. Mammalian Cell Maintenance and Transfection
3. Results
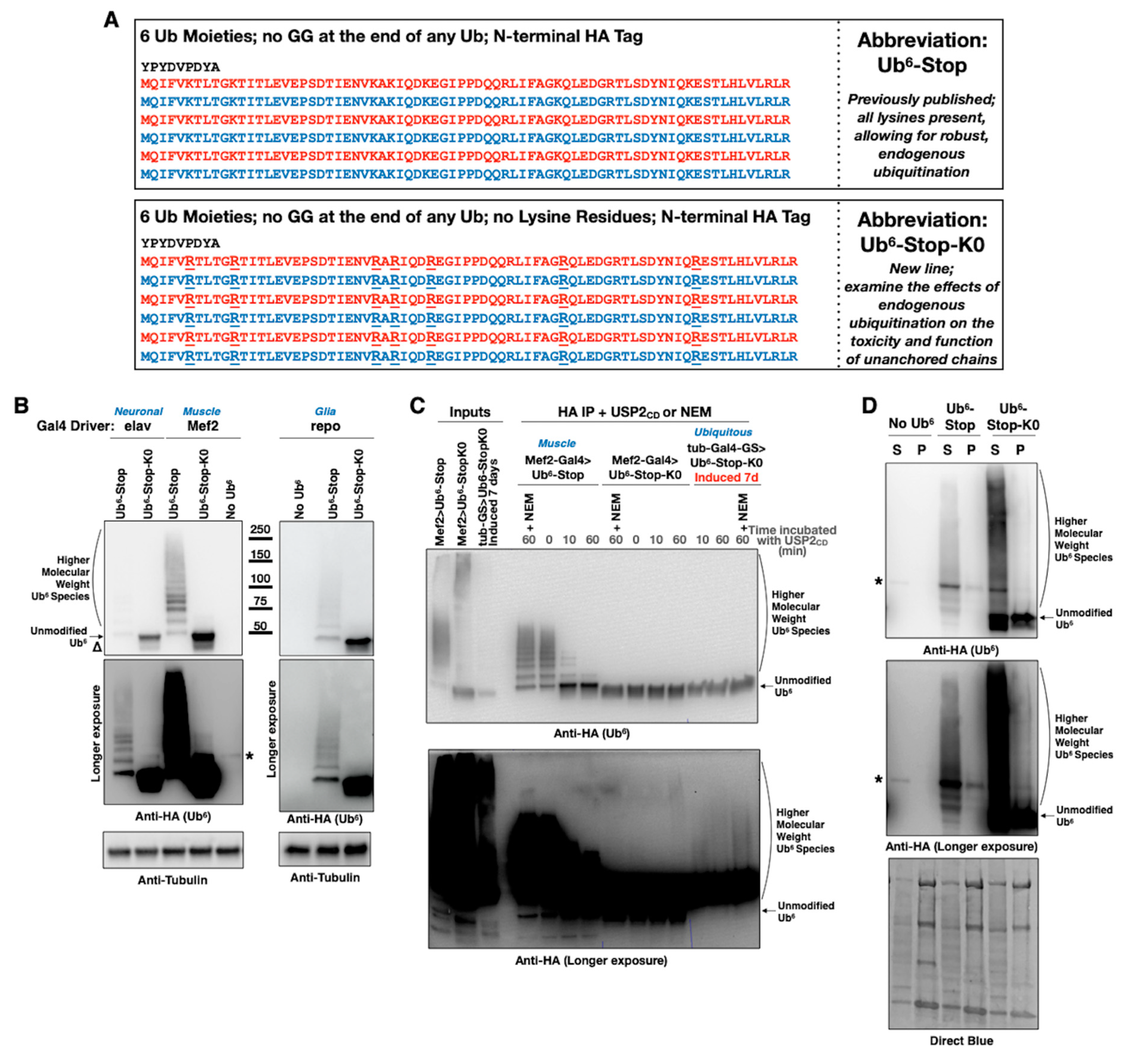
3.1. New Drosophila Lines Expressing Ubiquitination-Resistant, Unanchored Chains
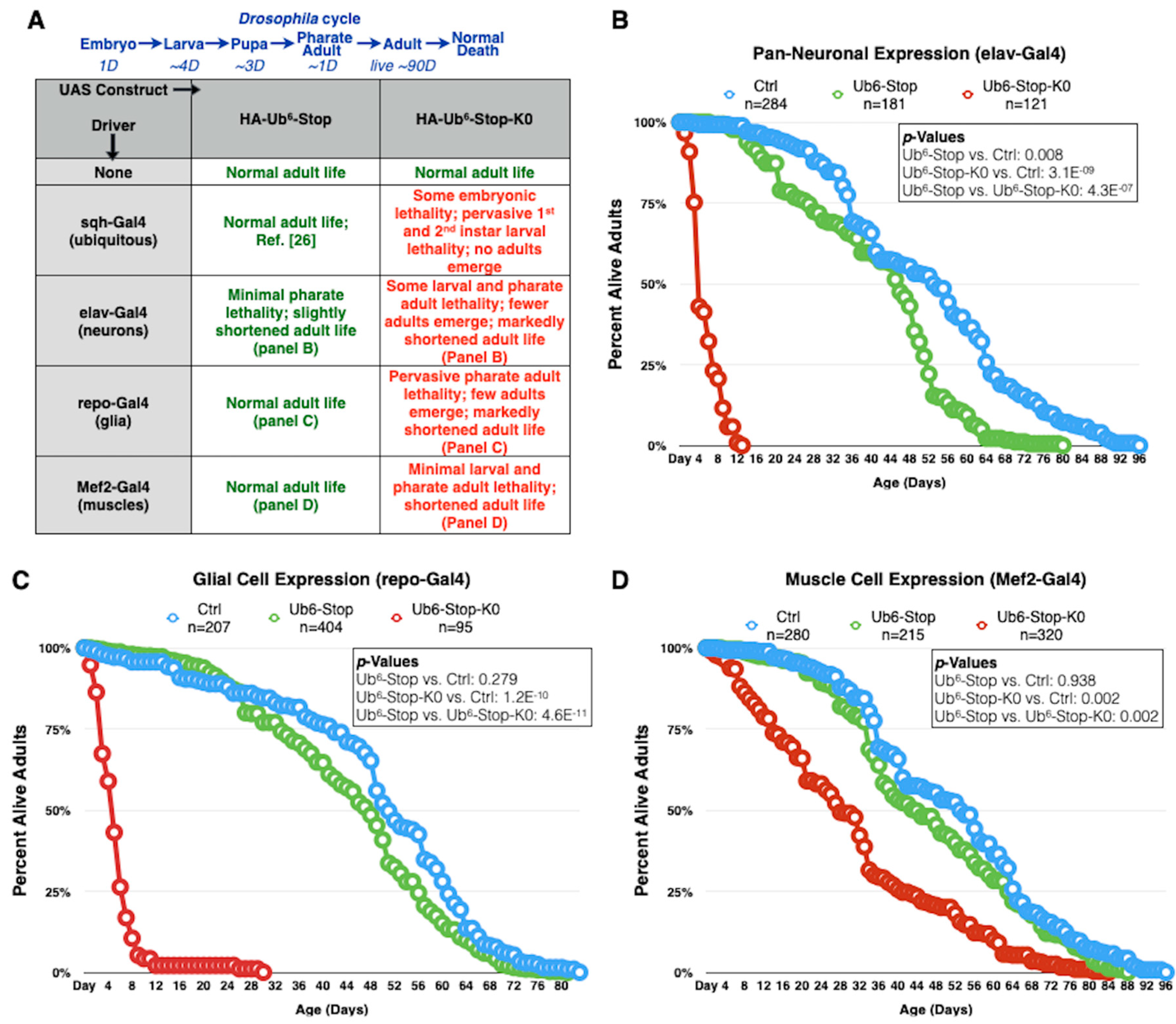
3.2. Lysine-Less, Unanchored Poly-Ub Is Highly Toxic in Drosophila
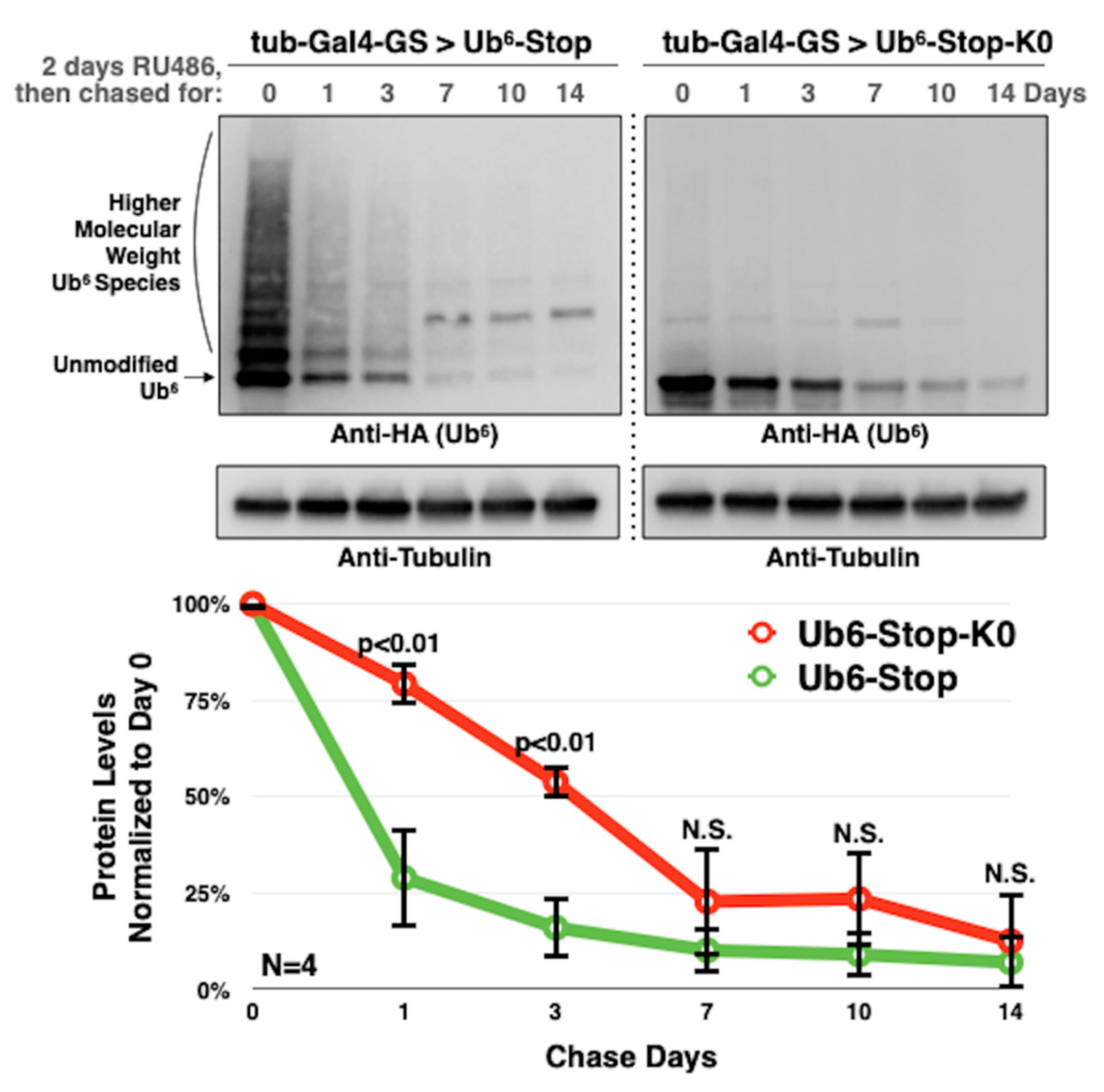
3.3. Lysine-Less, Unanchored Poly-Ub Is More Stable in the Fly
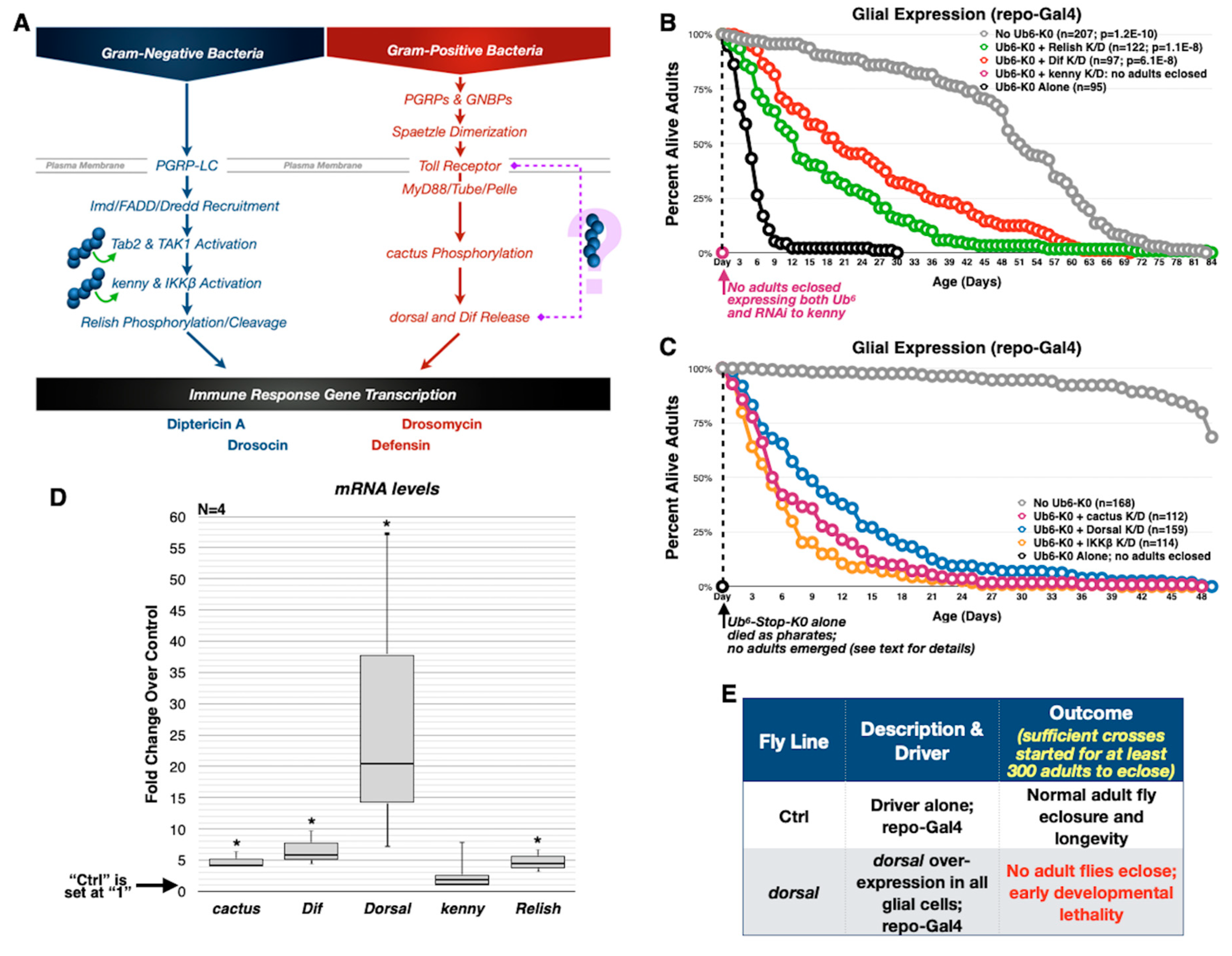
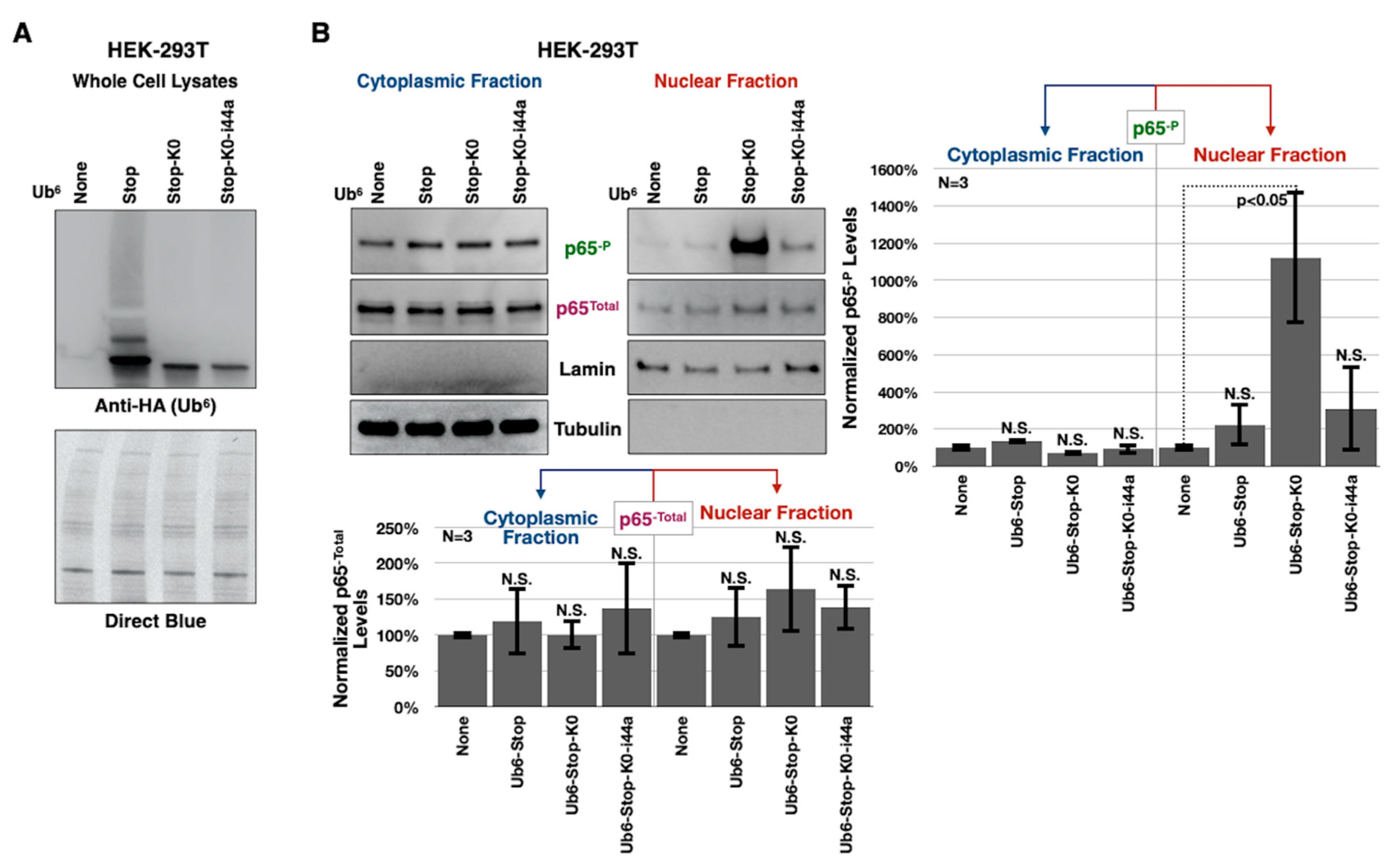
3.4. Toxicity of Lysine-Less, Unanchored Poly-Ub Depends on NF-κB Signaling
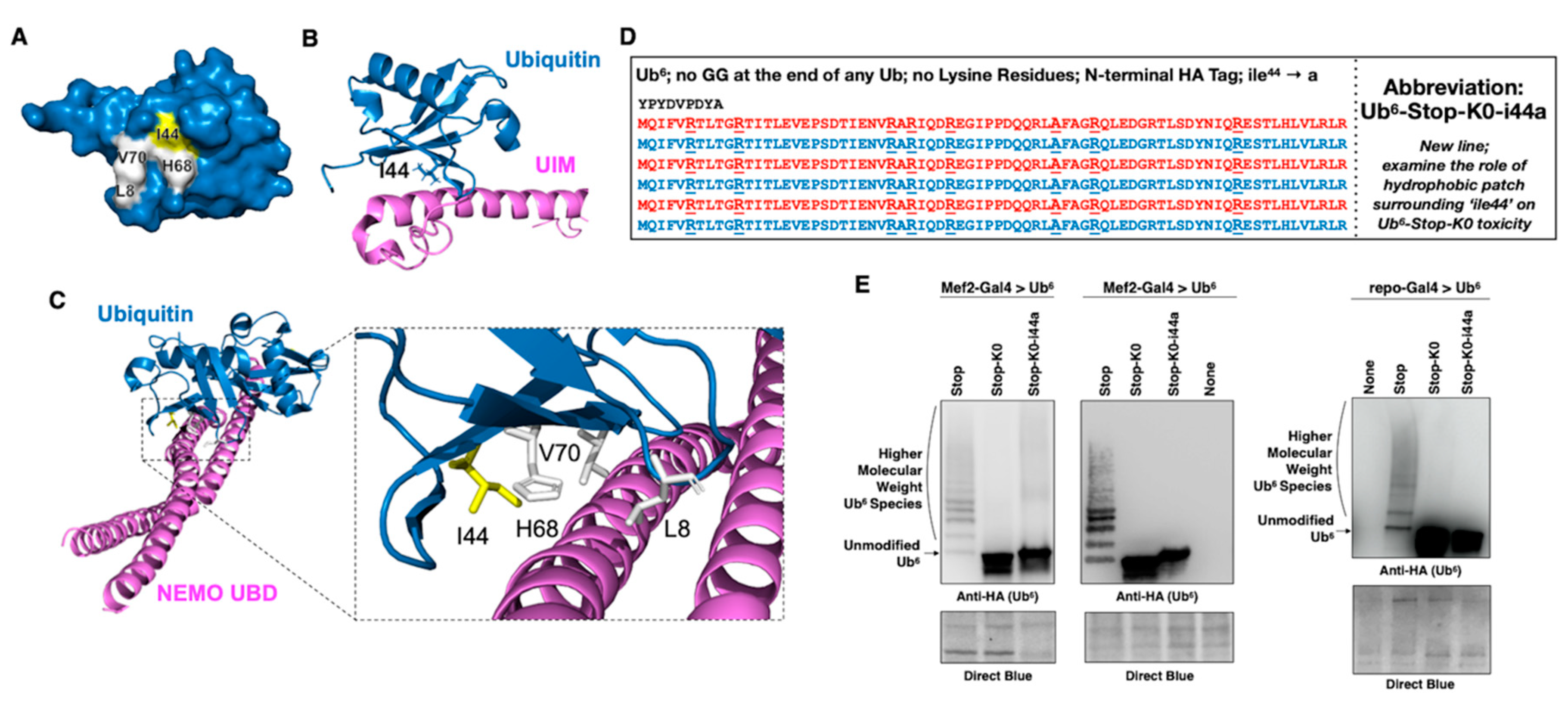
3.5. Aberrant NF-κB Signaling Is Mediated by the Ile44-Centered Hydrophobic Patch of Lysine-Less, Linear Poly-Ub
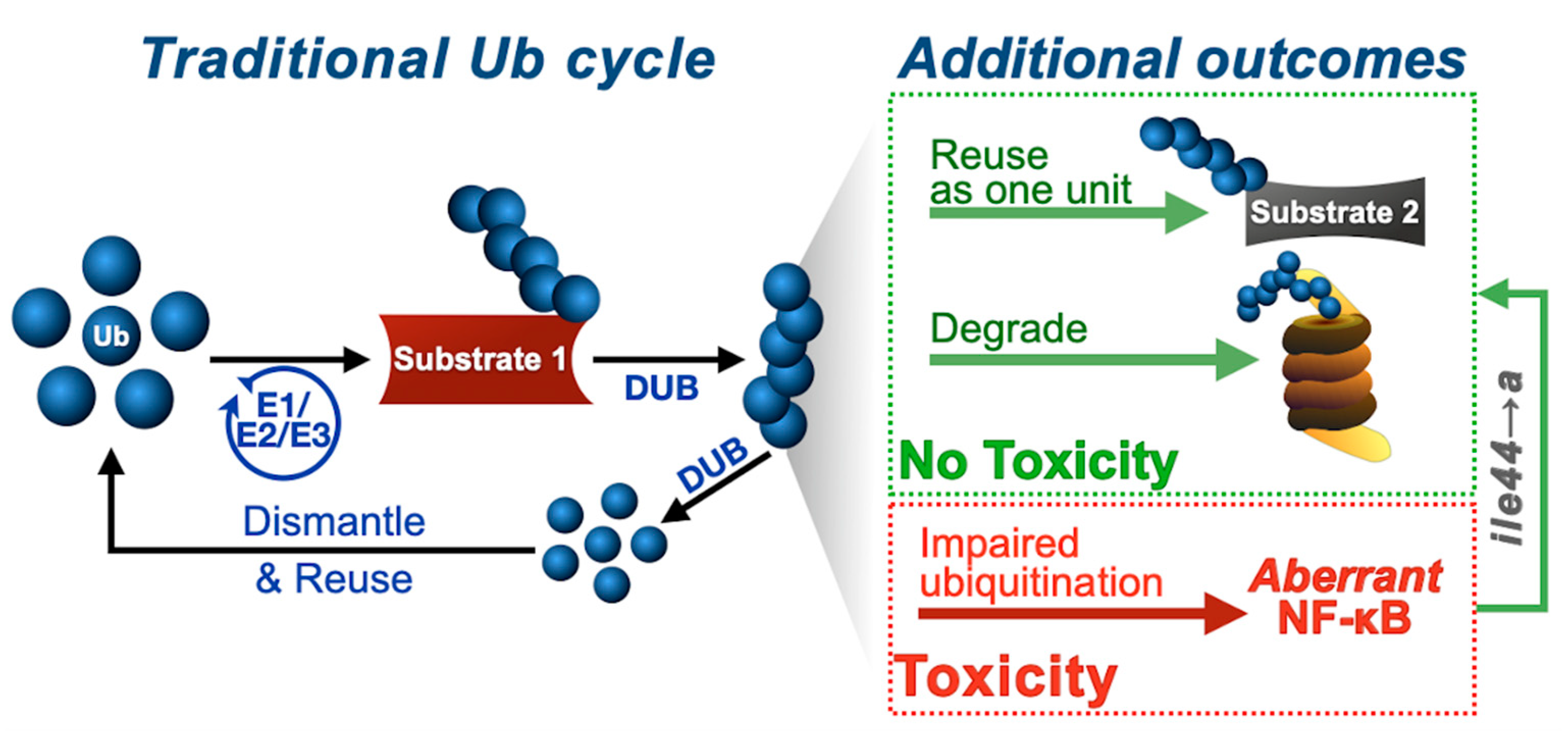
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stallcup, M.R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 2001, 20, 3014–3020. [Google Scholar] [CrossRef] [Green Version]
- Canagarajah, B.J.; Khokhlatchev, A.; Cobb, M.H.; Goldsmith, E.J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 1997, 90, 859–869. [Google Scholar] [CrossRef] [Green Version]
- Lis, H.; Sharon, N. Protein glycosylation. Structural and functional aspects. Eur. J. Biochem. 1993, 218, 1–27. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todi, S.V.; Scaglione, K.M.; Blount, J.R.; Basrur, V.; Conlon, K.P.; Pastore, A.; Elenitoba-Johnson, K.; Paulson, H.L. Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J. Biol. Chem. 2010, 285, 39303–39313. [Google Scholar] [CrossRef] [Green Version]
- Todi, S.V.; Winborn, B.J.; Scaglione, K.M.; Blount, J.R.; Travis, S.M.; Paulson, H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009, 28, 372–382. [Google Scholar] [CrossRef]
- Tsou, W.L.; Burr, A.A.; Ouyang, M.; Blount, J.R.; Scaglione, K.M.; Todi, S.V. Ubiquitination regulates the neuroprotective function of the deubiquitinase ataxin-3 in vivo. J. Biol. Chem. 2013, 288, 34460–34469. [Google Scholar] [CrossRef] [Green Version]
- Ristic, G.; Tsou, W.L.; Todi, S.V. An optimal ubiquitin-proteasome pathway in the nervous system: The role of deubiquitinating enzymes. Front. Mol. Neurosci. 2014, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.G.; Doerner, K.; Castellanos, E.R.; Haakonsen, D.L.; Werner, A.; Wang, N.; Yang, X.W.; Martinez-Martin, N.; Matsumoto, M.L.; Dixit, V.M.; et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933.e92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clague, M.J.; Coulson, J.M.; Urbe, S. Cellular functions of the DUBs. J. Cell Sci. 2012, 125, 277–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Shanks, J.R.; Komander, D.; Wilkinson, K.D. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 2008, 283, 19581–19592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmerich, C.H.; Ordureau, A.; Strickson, S.; Arthur, J.S.; Pedrioli, P.G.; Komander, D.; Cohen, P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. USA 2013, 110, 15247–15252. [Google Scholar] [CrossRef] [Green Version]
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.S.; Ito, A.; Yoshida, M.; Yao, T.P. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell. 2013, 51, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Braten, O.; Shabek, N.; Kravtsova-Ivantsiv, Y.; Ciechanover, A. Generation of free ubiquitin chains is up-regulated in stress and facilitated by the HECT domain ubiquitin ligases UFD4 and HUL5. Biochem. J. 2012, 444, 611–617. [Google Scholar] [CrossRef] [Green Version]
- Grou, C.P.; Pinto, M.P.; Mendes, A.V.; Domingues, P.; Azevedo, J.E. The de novo synthesis of ubiquitin: Identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 2015, 5, 12836. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, L.; Nagy, O.; Pal, M.; Udvardy, A.; Popescu, O.; Deak, P. Role of the deubiquitylating enzyme DmUsp5 in coupling ubiquitin equilibrium to development and apoptosis in Drosophila melanogaster. PLoS ONE 2015, 10, e0120875. [Google Scholar] [CrossRef] [Green Version]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from genes to organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Chen, G.C.; Chien, C.T. The deubiquitinase Leon/USP5 regulates ubiquitin homeostasis during Drosophila development. Biochem. Biophys. Res. Commun. 2014, 452, 369–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amerik, A.; Swaminathan, S.; Krantz, B.A.; Wilkinson, K.D.; Hochstrasser, M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997, 16, 4826–4838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowski, J.; Beal, R.; Hoffman, L.; Wilkinson, K.D.; Cohen, R.E.; Pickart, C.M. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 1997, 272, 23712–23721. [Google Scholar] [CrossRef] [Green Version]
- Dayal, S.; Sparks, A.; Jacob, J.; Allende-Vega, N.; Lane, D.P.; Saville, M.K. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 2009, 284, 5030–5041. [Google Scholar] [CrossRef] [Green Version]
- Blount, J.R.; Libohova, K.; Marsh, G.B.; Sutton, J.R.; Todi, S.V. Expression and Regulation of Deubiquitinase-Resistant, Unanchored Ubiquitin Chains in Drosophila. Sci. Rep. 2018, 8, 8513. [Google Scholar] [CrossRef]
- Blount, J.R.; Meyer, D.N.; Akemann, C.; Johnson, S.L.; Gurdziel, K.; Baker, T.R.; Todi, S.V. Unanchored ubiquitin chains do not lead to marked alterations in gene expression in Drosophila melanogaster. Biol. Open 2019, 8, bio043372. [Google Scholar] [CrossRef] [Green Version]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef] [Green Version]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Beal, R.; Deveraux, Q.; Xia, G.; Rechsteiner, M.; Pickart, C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. USA 1996, 93, 861–866. [Google Scholar] [CrossRef] [Green Version]
- Beal, R.E.; Toscano-Cantaffa, D.; Young, P.; Rechsteiner, M.; Pickart, C.M. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry 1998, 37, 2925–2934. [Google Scholar] [CrossRef] [PubMed]
- Sloper-Mould, K.E.; Jemc, J.C.; Pickart, C.M.; Hicke, L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001, 276, 30483–30489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fish, M.P.; Groth, A.C.; Calos, M.P.; Nusse, R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2007, 2, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.C.; Fish, M.; Nusse, R.; Calos, M.P. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 2004, 166, 1775–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsou, W.L.; Hosking, R.R.; Burr, A.A.; Sutton, J.R.; Ouyang, M.; Du, X.; Gomez, C.M.; Todi, S.V. DnaJ-1 and karyopherin alpha3 suppress degeneration in a new Drosophila model of Spinocerebellar Ataxia Type 6. Hum. Mol. Genet. 2015, 24, 4385–4396. [Google Scholar] [CrossRef] [Green Version]
- Tsou, W.L.; Qiblawi, S.H.; Hosking, R.R.; Gomez, C.M.; Todi, S.V. Polyglutamine length-dependent toxicity from alpha1ACT in Drosophila models of spinocerebellar ataxia type 6. Biol. Open 2016, 5, 1770–1775. [Google Scholar] [CrossRef] [PubMed]
- Sujkowski, A.; Bazzell, B.; Carpenter, K.; Arking, R.; Wessells, R.J. Endurance exercise and selective breeding for longevity extend Drosophila healthspan by overlapping mechanisms. Aging (Albany NY) 2015, 7, 535–552. [Google Scholar] [CrossRef] [Green Version]
- Brand, A.H.; Manoukian, A.S.; Perrimon, N. Ectopic expression in Drosophila. Methods Cell Biol. 1994, 44, 635–654. [Google Scholar]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993, 118, 401–415. [Google Scholar]
- Sutton, J.R.; Blount, J.R.; Libohova, K.; Tsou, W.L.; Joshi, G.S.; Paulson, H.L.; Costa, M.D.C.; Scaglione, K.M.; Todi, S.V. Interaction of the polyglutamine protein ataxin-3 with Rad23 regulates toxicity in Drosophila models of Spinocerebellar Ataxia Type 3. Hum. Mol. Genet. 2017, 26, 1419–1431. [Google Scholar] [CrossRef] [Green Version]
- Ristic, G.; Sutton, J.R.; Libohova, K.; Todi, S.V. Toxicity and aggregation of the polyglutamine disease protein, ataxin-3 is regulated by its binding to VCP/p97 in Drosophila melanogaster. Neurobiol. Dis. 2018, 116, 78–92. [Google Scholar] [CrossRef]
- Johnson, S.L.; Blount, J.R.; Libohova, K.; Ranxhi, B.; Paulson, H.L.; Tsou, W.L.; Todi, S.V. Differential toxicity of ataxin-3 isoforms in Drosophila models of Spinocerebellar Ataxia Type 3. Neurobiol. Dis. 2019, 132, 104535. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Elliott, P.R.; Pinto-Fernandez, A.; Bonham, S.; Kessler, B.M.; Komander, D.; El Oualid, F.; Krappmann, D. A Linear Diubiquitin-Based Probe for Efficient and Selective Detection of the Deubiquitinating Enzyme OTULIN. Cell Chem. Biol. 2017, 24, 1299–1313.e1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todi, S.V.; Franke, J.D.; Kiehart, D.P.; Eberl, D.F. Myosin VIIA defects, which underlie the Usher 1B syndrome in humans, lead to deafness in Drosophila. Curr. Biol. 2005, 15, 862–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, J.D.; Dong, F.; Rickoll, W.L.; Kelley, M.J.; Kiehart, D.P. Rod mutations associated with MYH9-related disorders disrupt nonmuscle myosin-IIA assembly. Blood 2005, 105, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Chen, Z.J. Regulation of NF-kappaB by ubiquitination. Curr. Opin. Immunol. 2013, 25, 4–12. [Google Scholar] [CrossRef]
- Courtois, G.; Fauvarque, M.O. The Many Roles of Ubiquitin in NF-kappaB Signaling. Biomedicines 2018, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.P.; Sun, L.; Chen, X.; Pineda, G.; Jiang, X.; Adhikari, A.; Zeng, W.; Chen, Z.J. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 2009, 461, 114–119. [Google Scholar] [CrossRef] [Green Version]
- Skaug, B.; Chen, J.; Du, F.; He, J.; Ma, A.; Chen, Z.J. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell. 2011, 44, 559–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, W.; Sun, L.; Jiang, X.; Chen, X.; Hou, F.; Adhikari, A.; Xu, M.; Chen, Z.J. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 2010, 141, 315–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catici, D.A.; Horne, J.E.; Cooper, G.E.; Pudney, C.R. Polyubiquitin Drives the Molecular Interactions of the NF-kappaB Essential Modulator (NEMO) by Allosteric Regulation. J. Biol. Chem. 2015, 290, 14130–14139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmar, G.; Winklhofer, K.F. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front. Chem. 2019, 7, 915. [Google Scholar] [CrossRef]
- Hicke, L.; Schubert, H.L.; Hill, C.P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005, 6, 610–621. [Google Scholar] [CrossRef]
- Laplantine, E.; Fontan, E.; Chiaravalli, J.; Lopez, T.; Lakisic, G.; Veron, M.; Agou, F.; Israel, A. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009, 28, 2885–2895. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshikawa, A.; Yamashita, M.; Yamagata, A.; Fukai, S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009, 28, 3903–3909. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, C.Y.; Barberi, T.J.; Ghosh, P.; Longo, D.L. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J. Biol. Chem. 2005, 280, 34538–34547. [Google Scholar] [CrossRef] [Green Version]
- Choe, K.M.; Werner, T.; Stoven, S.; Hultmark, D.; Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 2002, 296, 359–362. [Google Scholar] [CrossRef] [Green Version]
- Gottar, M.; Gobert, V.; Michel, T.; Belvin, M.; Duyk, G.; Hoffmann, J.A.; Ferrandon, D.; Royet, J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002, 416, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Ramet, M.; Manfruelli, P.; Pearson, A.; Mathey-Prevot, B.; Ezekowitz, R.A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 2002, 416, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wu, L.P.; Anderson, K.V. The antibacterial arm of the drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001, 15, 104–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutschmann, S.; Jung, A.C.; Zhou, R.; Silverman, N.; Hoffmann, J.A.; Ferrandon, D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat. Immunol. 2000, 1, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Silverman, N.; Zhou, R.; Stoven, S.; Pandey, N.; Hultmark, D.; Maniatis, T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000, 14, 2461–2471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoven, S.; Ando, I.; Kadalayil, L.; Engstrom, Y.; Hultmark, D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000, 1, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Stoven, S.; Silverman, N.; Junell, A.; Hedengren-Olcott, M.; Erturk, D.; Engstrom, Y.; Maniatis, T.; Hultmark, D. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc. Natl. Acad. Sci. USA 2003, 100, 5991–5996. [Google Scholar] [CrossRef] [Green Version]
- Myllymaki, H.; Valanne, S.; Ramet, M. The Drosophila imd signaling pathway. J. Immunol. 2014, 192, 3455–3462. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Pickart, C.M. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J. Biol. Chem. 1990, 265, 21835–21842. [Google Scholar]
- Park, E.S.; Elangovan, M.; Kim, Y.J.; Yoo, Y.J. UbcD4, an ortholog of E2-25K/Ube2K, is essential for activation of the immune deficiency pathway in Drosophila. Biochem. Biophys. Res. Commun. 2016, 469, 891–896. [Google Scholar] [CrossRef]
- Zhou, R.; Silverman, N.; Hong, M.; Liao, D.S.; Chung, Y.; Chen, Z.J.; Maniatis, T. The role of ubiquitination in Drosophila innate immunity. J. Biol. Chem. 2005, 280, 34048–34055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Z.; Li, Z.; Xiao, W. Drosophila bendless catalyzes K63-linked polyubiquitination and is involved in the response to DNA damage. Mutat. Res. 2018, 808, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Arnot, C.J.; Gay, N.J.; Gangloff, M. Molecular mechanism that induces activation of Spatzle, the ligand for the Drosophila Toll receptor. J. Biol. Chem. 2010, 285, 19502–19509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangloff, M.; Murali, A.; Xiong, J.; Arnot, C.J.; Weber, A.N.; Sandercock, A.M.; Robinson, C.V.; Sarisky, R.; Holzenburg, A.; Kao, C.; et al. Structural insight into the mechanism of activation of the Toll receptor by the dimeric ligand Spatzle. J. Biol. Chem. 2008, 283, 14629–14635. [Google Scholar] [CrossRef] [Green Version]
- Weber, A.N.; Moncrieffe, M.C.; Gangloff, M.; Imler, J.L.; Gay, N.J. Ligand-receptor and receptor-receptor interactions act in concert to activate signaling in the Drosophila toll pathway. J. Biol. Chem. 2005, 280, 22793–22799. [Google Scholar] [CrossRef] [Green Version]
- Towb, P.; Bergmann, A.; Wasserman, S.A. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development 2001, 128, 4729–4736. [Google Scholar]
- Wu, L.P.; Anderson, K.V. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature 1998, 392, 93–97. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.H.; Ramet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Model Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blount, J.R.; Libohova, K.; Silva, G.M.; Todi, S.V. Isoleucine 44 Hydrophobic Patch Controls Toxicity of Unanchored, Linear Ubiquitin Chains through NF-κB Signaling. Cells 2020, 9, 1519. https://doi.org/10.3390/cells9061519
Blount JR, Libohova K, Silva GM, Todi SV. Isoleucine 44 Hydrophobic Patch Controls Toxicity of Unanchored, Linear Ubiquitin Chains through NF-κB Signaling. Cells. 2020; 9(6):1519. https://doi.org/10.3390/cells9061519
Chicago/Turabian StyleBlount, Jessica R., Kozeta Libohova, Gustavo M. Silva, and Sokol V. Todi. 2020. "Isoleucine 44 Hydrophobic Patch Controls Toxicity of Unanchored, Linear Ubiquitin Chains through NF-κB Signaling" Cells 9, no. 6: 1519. https://doi.org/10.3390/cells9061519
APA StyleBlount, J. R., Libohova, K., Silva, G. M., & Todi, S. V. (2020). Isoleucine 44 Hydrophobic Patch Controls Toxicity of Unanchored, Linear Ubiquitin Chains through NF-κB Signaling. Cells, 9(6), 1519. https://doi.org/10.3390/cells9061519




