Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance
Abstract
1. Introduction
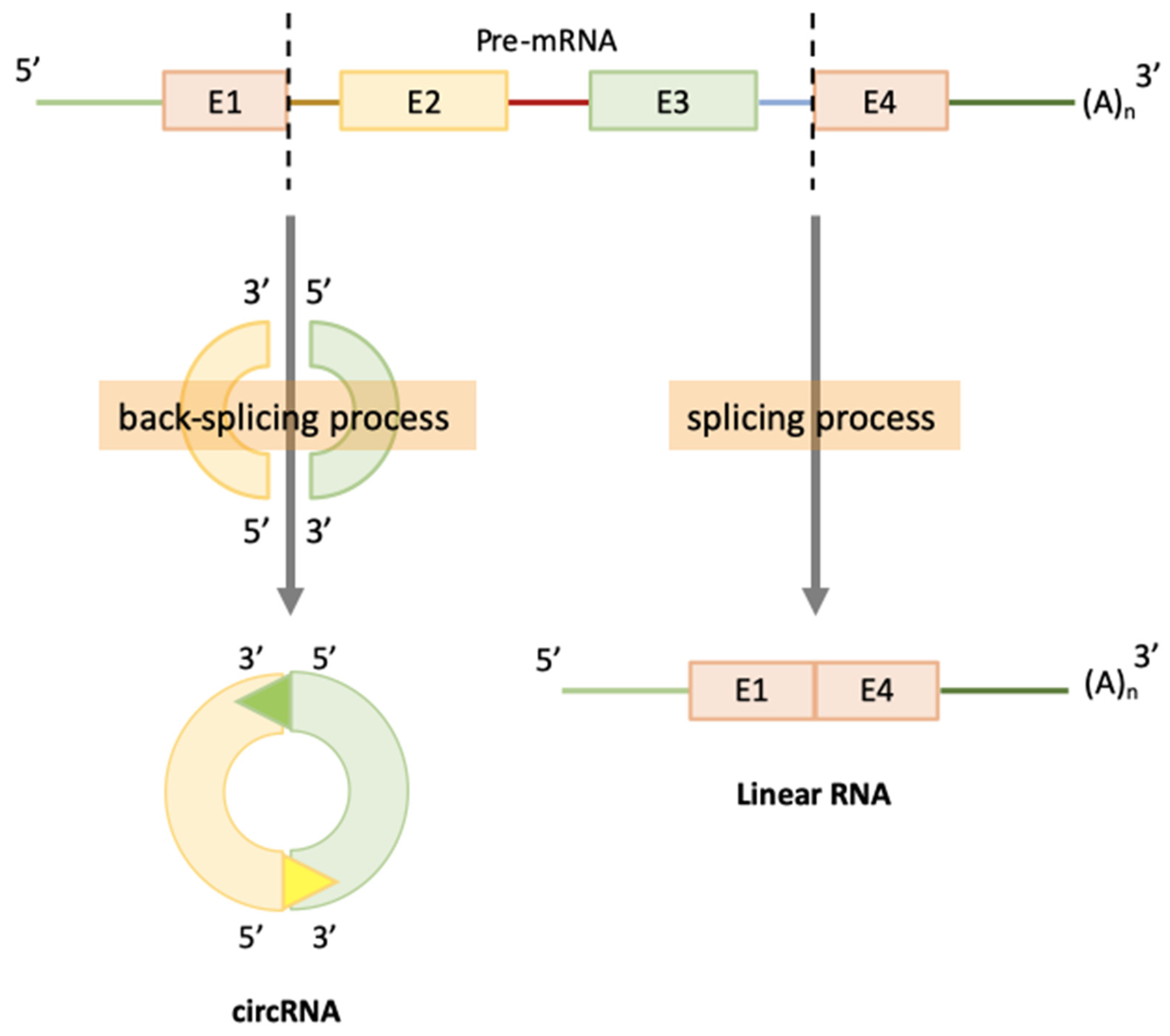
2. CircRNAs Biogenesis
3. Technologies Available to Analyze CircRNAs as Biomarkers
4. CircRNAs Function in Sepsis
4.1. Role of CircRNAs in Inflammation
4.2. Role of CircRNAs in Immunosuppression
4.3. Role of CircRNAs in Endothelium Dysfunction
5. Clinical Significance
Funding
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock. JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the Global Burden of Disease Study. Lancet (Lond. UK) 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Reinhart, K.; Daniels, R.; Kissoon, N.; Machado, F.R.; Schachter, R.D.; Finfer, S. Recognizing sepsis as a Global Health Priority — A WHO resolution. N. Engl. J. Med. 2017, 377, 414–417. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020. [Google Scholar] [CrossRef]
- Tg Experts discuss link between sepsis and Covid-19. Available online: https://european-biotechnology.com/up-to-date/latest-news/news/experts-discuss-link-between-sepsis-and-covid-19.html (accessed on 25 March 2020).
- Pierrakos, C.; Vincent, J.L. Sepsis biomarkers: A review. Crit. Care 2010, 14, 1–18. [Google Scholar] [CrossRef]
- Jensen, J.U.; Bouadma, L. Why biomarkers failed in sepsis. Intensive Care Med. 2016, 42, 2049–2051. [Google Scholar] [CrossRef]
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and costs of sepsis in the United States-An analysis based on timing of diagnosis and severity level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, T.; Xiao, J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018, 34, 267–274. [Google Scholar] [CrossRef]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.-M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Dong, R.; Zhang, Y.; Zhang, J.-L.; Luo, Z.; Zhang, J.; Chen, L.-L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Yang, J.; Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 2020, 11, 90. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genomics 2017, 2017, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer – Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhou, L.; Ponnusamy, M.; Zhang, L.; Dong, Y.; Zhang, Y.; Wang, Q.; Liu, J.; Wang, K. A comprehensive review of circRNA: From purification and identification to disease marker potential. PeerJ 2018, 6, e5503. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Hanan, M.; Soreq, H.; Kadener, S. CircRNAs in the brain. RNA Biol. 2017, 14, 1028–1034. [Google Scholar] [CrossRef]
- Gruner, H.; Cortés-López, M.; Cooper, D.A.; Bauer, M.; Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016, 6, 38907. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Dong, R.; Ma, X.-K.; Chen, L.-L.; Yang, L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017, 14, 1064–1074. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Morillon, A. Definition and Families of Long Non-coding RNA. In Long Non-Coding RNA; Elsevier: Amsterdam, The Netherlands, 2018; pp. 25–53. [Google Scholar]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1, 67–79. [Google Scholar]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Ebbesen, K.K.; Kjems, J.; Hansen, T.B. Circular RNAs: Identification, biogenesis and function. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ding, W.; Sun, T.; Tariq, M.A.; Xu, T.; Li, P.; Wang, J. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018, 285, 220–232. [Google Scholar] [CrossRef]
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, K.; Wu, F.; Wang, W.; Zhang, K.; Hu, H.; Liu, Y.; Jiang, T. circRNA disease: A manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018, 9, 475. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Li, D.; Xia, J.; Wu, Q.-J.; Wen, R.; Yang, N.; Liu, C.-F. Non-coding RNA: A potential biomarker and therapeutic target for sepsis. Oncotarget 2017, 8, 91765–91778. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–561. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef]
- Xie, L.; Mao, M.; Xiong, K.; Jiang, B. Circular RNAs: A Novel Player in Development and Disease of the Central Nervous System. Front. Cell. Neurosci. 2017, 11, 354. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef]
- Diallo, L.H.; Tatin, F.; David, F.; Godet, A.-C.; Zamora, A.; Prats, A.-C.; Garmy-Susini, B.; Lacazette, E. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie 2019, 164, 45–52. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef]
- Chang, C.-C.; Huang, R.-L.; Liao, Y.-P.; Su, P.-H.; Hsu, Y.-W.; Wang, H.-C.; Tien, C.-Y.; Yu, M.-H.; Lin, Y.-W.; Lai, H.-C. Concordance analysis of methylation biomarkers detection in self-collected and physician-collected samples in cervical neoplasm. BMC Cancer 2015, 15, 418. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Monaghan, S.F.; Banerjee, D.; Chung, C.-S.; Lomas-Neira, J.; Cygan, K.J.; Rhine, C.L.; Fairbrother, W.G.; Heffernan, D.S.; Levy, M.M.; Cioffi, W.G.; et al. Changes in the process of alternative RNA splicing results in soluble B and T lymphocyte attenuator with biological and clinical implications in critical illness. Mol. Med. 2018, 24, 32. [Google Scholar] [CrossRef]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2018, 75, 1071–1098. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016, 7, 120–160. [Google Scholar] [CrossRef]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef]
- Yang, L.; Fu, J.; Zhou, Y. Circular RNAs and Their Emerging Roles in Immune Regulation. Front. Immunol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.A.; Rowley, J.W.; Campbell, R.A.; Grissom, C.K.; Brown, S.M.; Beesley, S.J.; Schwertz, H.; Kosaka, Y.; Manne, B.K.; Krauel, K.; et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood 2019, 134, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Pandya-Jones, A.; Black, D.L. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009, 15, 1896–1908. [Google Scholar] [CrossRef]
- Brugiolo, M.; Herzel, L.; Neugebauer, K.M. Counting on co-transcriptional splicing. F1000Prime Rep. 2013, 5. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.-L.; Yang, L.; Chen, L.-L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef]
- Brzyżek, G.; Świeżewski, S. Mutual interdependence of splicing and transcription elongation. Transcription 2015, 6, 37–39. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Roy, C.K.; Olson, S.; Graveley, B.R.; Zamore, P.D.; Moore, M.J. Assessing long-distance RNA sequence connectivity via RNA-templated DNA–DNA ligation. Elife 2015, 4, 1–13. [Google Scholar] [CrossRef]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef]
- Danan, M.; Schwartz, S.; Edelheit, S.; Sorek, R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012, 40, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Lv, Y.; Liang, X.; Zhao, B.; Bian, W.; Zhang, D.; Jiang, J.; Zhang, C. Circular RNA Expression Profiles in Plasma from Patients with Heart Failure Related to Platelet Activity. Biomolecules 2020, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Mills, J.D.; Takenaka, K.; Bliim, N.; Halliday, G.M.; Janitz, M. Characterization of circular RNAs landscape in multiple system atrophy brain. J. Neurochem. 2016, 139, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Venø, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Lin, W.; Guo, M.; Zou, Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017, 13, e1005420. [Google Scholar] [CrossRef]
- Meng, X.; Hu, D.; Zhang, P.; Chen, Q.; Chen, M. CircFunBase: A database for functional circular RNAs. Database 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Kingsley, S.M.K.; Bhat, B.V. Role of microRNAs in sepsis. Inflamm. Res. 2017, 66, 553–569. [Google Scholar] [CrossRef]
- Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating microRNAs as biomarkers for sepsis. Int. J. Mol. Sci. 2016, 17, 78. [Google Scholar] [CrossRef]
- Qin, L.; Lin, J.; Xie, X. CircRNA-9119 suppresses poly I:C induced inflammation in Leydig and Sertoli cells via TLR3 and RIG-I signal pathways. Mol. Med. 2019, 25, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-C.; Guo, X.-L.; Li, X. The novel roles of circular RNAs in metabolic organs. Genes Dis. 2018, 5, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, C.-X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.-F.; Wei, J.; Yao, R.-W.; Yang, L.; Chen, L.-L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Chousterman, B.G.; Swirski, F.K.; Weber, G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017, 39, 517–528. [Google Scholar] [CrossRef]
- Kumar, V. Immunometabolism: Another Road to Sepsis and Its Therapeutic Targeting. Inflammation 2019, 42, 765–788. [Google Scholar] [CrossRef]
- Dan, C.; Jinjun, B.; Zi-Chun, H.; Lin, M.; Wei, C.; Xu, Z.; Ri, Z.; Shun, C.; Wen-Zhu, S.; Qing-Cai, J.; et al. Modulation of TNF-α mRNA stability by human antigen R and miR181s in sepsis-induced immunoparalysis. EMBO Mol. Med. 2015, 7, 140–157. [Google Scholar] [CrossRef]
- Huang, H.-C.; Yu, H.-R.; Huang, L.-T.; Huang, H.-C.; Chen, R.-F.; Lin, I.-C.; Ou, C.-Y.; Hsu, T.-Y.; Yang, K.D. miRNA-125b regulates TNF-α production in CD14+ neonatal monocytes via post-transcriptional regulation. J. Leukoc. Biol. 2012, 92, 171–182. [Google Scholar] [CrossRef]
- Puimège, L.; Van Hauwermeiren, F.; Steeland, S.; Van Ryckeghem, S.; Vandewalle, J.; Lodens, S.; Dejager, L.; Vandevyver, S.; Staelens, J.; Timmermans, S.; et al. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol. Med. 2015, 7, 1004–1017. [Google Scholar] [CrossRef]
- Mera, S.; Tatulescu, D.; Cismaru, C.; Bondor, C.; Slavcovici, A.; Zanc, V.; Carstina, D.; Oltean, M. Multiplex cytokine profiling in patients with sepsis. APMIS 2011, 119, 155–163. [Google Scholar] [CrossRef]
- Zhou, J.; Chaudhry, H.; Zhong, Y.; Ali, M.M.; Perkins, L.A.; Owens, W.B.; Morales, J.E.; McGuire, F.R.; Zumbrun, E.E.; Zhang, J.; et al. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine 2015, 71, 89–100. [Google Scholar] [CrossRef]
- Gao, M.; Wang, X.; Zhang, X.; Ha, T.; Ma, H.; Liu, L.; Kalbfleisch, J.H.; Gao, X.; Kao, R.L.; Williams, D.L.; et al. Attenuation of Cardiac Dysfunction in Polymicrobial Sepsis by MicroRNA-146a Is Mediated via Targeting of IRAK1 and TRAF6 Expression. J. Immunol. 2015, 195, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ghosh, S. Toll-like receptor-mediated NF-kappaB activation: A phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 2001, 107, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Grobmyer, S.R.; Lin, E.; Lowry, S.F.; Rivadeneira, D.E.; Potter, S.; Barie, P.S.; Nathan, C.F. Elevation of IL-18 in human sepsis. J. Clin. Immunol. 2000, 20, 212–215. [Google Scholar] [CrossRef]
- Endo, S.; Inada, K.; Yamada, Y.; Wakabayashi, G.; Ishikura, H.; Tanaka, T.; Sato, S. Interleukin 18 (IL-18) levels in patients with sepsis. J. Med. 2000, 31, 15–20. [Google Scholar]
- Guzzo, C.; Ayer, A.; Basta, S.; Banfield, B.W.; Gee, K. IL-27 Enhances LPS-Induced Proinflammatory Cytokine Production via Upregulation of TLR4 Expression and Signaling in Human Monocytes. J. Immunol. 2012, 188, 864–873. [Google Scholar] [CrossRef]
- Pflanz, S.; Timans, J.C.; Cheung, J.; Rosales, R.; Kanzler, H.; Gilbert, J.; Hibbert, L.; Churakova, T.; Travis, M.; Vaisberg, E.; et al. IL-27, a Heterodimeric Cytokine Composed of EBI3 and p28 Protein, Induces Proliferation of Naive CD4+ T Cells. Immunity 2002, 16, 779–790. [Google Scholar] [CrossRef]
- Wang, H.; He, P.; Pan, H.; Long, J.; Wang, J.; Li, Z.; Liu, H.; Jiang, W.; Zheng, Z. Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 2018, 50, 27. [Google Scholar] [CrossRef]
- Sheng, Y.-J.; Gao, J.-P.; Li, J.; Han, J.-W.; Xu, Q.; Hu, W.-L.; Pan, T.-M.; Cheng, Y.-L.; Yu, Z.-Y.; Ni, C.; et al. Follow-up study identifies two novel susceptibility loci PRKCB and 8p11.21 for systemic lupus erythematosus. Rheumatology (Oxford) 2011, 50, 682–688. [Google Scholar] [CrossRef]
- Deng, T.; Yang, L.; Zheng, Z.; Li, Y.; Ren, W.; Wu, C.; Guo, L. Calcitonin gene-related peptide induces IL-6 expression in RAW264.7 macrophages mediated by mmu_circRNA_007893. Mol. Med. Rep. 2017, 16, 9367–9374. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zhang, Y.; Wang, J.-J. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol. Int. 2017, 41, 1283–1289. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Chen, Y.; Wu, Z.; Zhang, C.; Shi, W. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4+ T cells of systemic lupus erythematous. Clin. Sci. (Lond.) 2018, 132, 2285–2298. [Google Scholar] [CrossRef] [PubMed]
- Gentile, L.F.; Cuenca, A.G.; Efron, P.A.; Ang, D.; Bihorac, A.; McKinley, B.A.; Moldawer, L.L.; Moore, F.A. Persistent inflammation and immunosuppression: A common syndrome and new horizon for surgical intensive care. J. Trauma Acute Care Surg. 2012, 72, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Delano, M.J.; Scumpia, P.O.; Weinstein, J.S.; Coco, D.; Nagaraj, S.; Kelly-Scumpia, K.M.; O’Malley, K.A.; Wynn, J.L.; Antonenko, S.; Al-Quran, S.Z.; et al. MyD88-dependent expansion of an immature GR-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J. Exp. Med. 2007, 204, 1463–1474. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.J.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Rajewsky, K. MicroRNA control in the immune system: BBasic principles. Cell 2009, 136, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.-T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Niu, P.; Zhao, Y.; Cheng, Y.; Chen, W.; Lin, L.; Lu, J.; Cheng, X.; Xu, Z. Impact of miR-223-3p and miR-2909 on inflammatory factors IL-6, IL-1ß, and TNF-α, and the TLR4/TLR2/NF-κB/STAT3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PLoS ONE 2019, 14, e0212063. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lv, W.; Lin, Z.; Wang, X.; Bu, J.; Su, Y. Hsa_circ_0003159 inhibits gastric cancer progression by regulating miR-223-3p/NDRG1 axis. Cancer Cell Int. 2020, 20, 57. [Google Scholar] [CrossRef]
- Wang, J.; Yu, M.; Yu, G.; Bian, J.; Deng, X.; Wan, X.; Zhu, K. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, Y.; Jiang, Y. The prognostic value of plasma microRNA-155 and microRNA-146a level in severe sepsis and sepsis-induced acute lung injury patients. Clin. Lab. 2016, 62, 2355–2360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, B.; Chen, G.; Zhang, L.; Zhuang, Y.; Niu, H.; Zeng, Z. Circular RNA RSF1 promotes inflammatory and fibrotic phenotypes of irradiated hepatic stellate cell by modulating miR-146a-5p. J. Cell. Physiol. 2020, 1, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.Q.; Cai, C.W.; Shen, J.; Zheng, Q.; Ran, Z.H. Circular RNA expression alterations in colon tissues of Crohn’s disease patients. Mol. Med. Rep. 2019, 19, 4500–4506. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X. MiR-125b but not miR-125a is upregulated and exhibits a trend to correlate with enhanced disease severity, inflammation, and increased mortality in sepsis patients. J. Clin. Lab. Anal. 2020, 34, 1–12. [Google Scholar] [CrossRef]
- Li, J.-F.; Song, Y.-Z. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumor Biol. 2017, 39, 100–104. [Google Scholar] [CrossRef]
- Caserta, S.; Kern, F.; Cohen, J.; Drage, S.; Newbury, S.F.; Llewelyn, M.J. Circulating plasma microRNAs can differentiate human sepsis and Systemic Inflammatory Response Syndrome (SIRS). Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, X.; Hu, X.; Yuan, L.; Cheng, J.; Jiang, Y.; Ao, Y. Emerging Roles of circRNA Related to the Mechanical Stress in Human Cartilage Degradation of Osteoarthritis. Mol. Ther. Nucleic Acids 2017, 7, 223–230. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, Z.; Mao, Y.; Dong, W.; Zhang, Y.; Yin, N.; Jiang, L. miR-27a is up regulated and promotes inflammatory response in sepsis. Cell. Immunol. 2014, 290, 190–195. [Google Scholar] [CrossRef]
- Liu, S.; Liu, C.; Wang, Z.; Huang, J.; Zeng, Q. microRNA-23a-5p acts as a potential biomarker for sepsis-induced acute respiratory distress syndrome in early stage. Cell. Mol. Biol. 2016, 62, 31–37. [Google Scholar] [PubMed]
- Zhang, H.; Li, H.; Shaikh, A.; Caudle, Y.; Yao, B.; Yin, D. Inhibition of microRNA-23b attenuates immunosuppression during late sepsis through NIK, TRAF1 and XIAP. J. Infect. Dis. 2018, 218, 300–311. [Google Scholar] [CrossRef] [PubMed]
- García-Giménez, J.L.; Mena-Mollá, S.; Beltrán-García, J.; Sanchis-Gomar, F. Challenges in the analysis of epigenetic biomarkers in clinical samples. Clin. Chem. Lab. Med. 2017, 55, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
- Agirre, X.; Meydan, C.; Jiang, Y.; Garate, L.; Doane, A.S.; Li, Z.; Verma, A.; Paiva, B.; Martín-Subero, J.I.; Elemento, O.; et al. Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat. Commun. 2019, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- McClure, C.; Brudecki, L.; Ferguson, D.A.; Yao, Z.Q.; Moorman, J.P.; McCall, C.E.; El Gazzar, M. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect. Immun. 2014, 82, 3816–3825. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wang, T.; Tang, H.; Lv, Z.; Liang, P. Circular RNA circMAN2B2 facilitates glioma progression by regulating the miR-1205/S100A8 axis. J. Cell. Physiol. 2019, 234, 22996–23004. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, J.E.; Feng, Y.; Velazquez, H.; Sessa, W.C. Endothelial glucocorticoid receptor is required for protection against sepsis. Proc. Natl. Acad. Sci. USA 2013, 110, 306–311. [Google Scholar] [CrossRef]
- Stöcklin, E.; Wissler, M.; Gouilleux, F.; Groner, B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 1996, 383, 726–728. [Google Scholar] [CrossRef]
- Chow, J.C.; Ling, P.R.; Qu, Z.; Laviola, L.; Ciccarone, A.; Bistrian, B.R.; Smith, R.J. Growth hormone stimulates tyrosine phosphorylation of JAK2 and STAT5, but not insulin receptor substrate-1 or SHC proteins in liver and skeletal muscle of normal rats in vivo. Endocrinology 1996, 137, 2880–2886. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, Y.; Hou, J.; Shu, Q.; Yin, Y.; Fu, W.; Han, F.; Hou, T.; Zeng, C.; Nemeth, E.; et al. Increased gene copy number of DEFA1/DEFA3 worsens sepsis by inducing endothelial pyroptosis. Proc. Natl. Acad. Sci. USA 2019, 116, 3161–3170. [Google Scholar] [CrossRef]
- Ibañez-Cabellos, J.S.; Aguado, C.; Pérez-Cremades, D.; García-Giménez, J.L.; Bueno-Betí, C.; García-López, E.M.; Romá-Mateo, C.; Novella, S.; Hermenegildo, C.; Pallardó, F.V. Extracellular histones activate autophagy and apoptosis via mTOR signaling in human endothelial cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3234–3246. [Google Scholar] [CrossRef] [PubMed]
- Opitz, B.; Eitel, J.; Meixenberger, K.; Suttorp, N. Role of Toll-like receptors, NOD-like receptors and RIG-I-like receptors in endothelial cells and systemic infections. Thromb. Haemost. 2009, 102, 1103–1109. [Google Scholar] [PubMed]
- Elliott, E.I.; Sutterwala, F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015, 265, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, X.; Hu, X.; Dai, L.; Fu, X.; Zhang, J.; Ao, Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 “Sponge” in Human Cartilage Degradation. Sci. Rep. 2016, 6, 22572. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Yang, N.; Goodwin, J.E.; Mahrer, K.; Li, D.; Xia, J.; Wen, R.; Zhou, H.; Zhang, T.; Song, W.-L.; et al. Characterization of Circular RNA and microRNA Profiles in Septic Myocardial Depression: A Lipopolysaccharide-Induced Rat Septic Shock Model. Inflammation 2019, 42, 1990–2002. [Google Scholar] [CrossRef]
- Ng, W.L.; Marinov, G.K.; Liau, E.S.; Lam, Y.L.; Lim, Y.-Y.; Ea, C.-K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016, 13, 861–871. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, 92–97. [Google Scholar] [CrossRef]
- Xie, H.; Ren, X.; Xin, S.; Lan, X.; Lu, G.; Lin, Y.; Yang, S.; Zeng, Z.; Liao, W.; Ding, Y.-Q.; et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016, 7, 26680–26691. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 124–129. [Google Scholar] [CrossRef]
- Chen, L.-Y.; Zhi, Z.; Wang, L.; Zhao, Y.-Y.; Deng, M.; Liu, Y.-H.; Qin, Y.; Tian, M.-M.; Liu, Y.; Shen, T.; et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J. Pathol. 2019, 248, 103–115. [Google Scholar] [CrossRef]
- Li, P.; Chen, H.; Chen, S.; Mo, X.; Li, T.; Xiao, B.; Yu, R.; Guo, J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 2017, 116, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA circHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging miR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Wang, T.; Ge, Q.; Xu, H.; Wu, Y.; Tang, Q.; Chen, K. Circular RNA Signature in Hepatocellular Carcinoma. J. Cancer 2019, 10, 3361–3372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Che, S.; Cui, J.; Liu, Y.; An, X.; Cao, B.; Song, Y. CircRNA-9119 regulates the expression of prostaglandin-endoperoxide synthase 2 (PTGS2) by sponging miR-26a in the endometrial epithelial cells of dairy goat. Reprod. Fertil. Dev. 2018, 30, 1759–1769. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Wei, C.-Y.; Huang, X.-Y.; Peng, R.; Yang, X.; Lu, J.-C.; Zhang, C.; Gao, C.; Cai, J.-B.; Gao, P.-T.; et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 2019, 18, 105. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Mira, J.-P.; Antonelli, M. Sepsis: Older and newer concepts. Lancet. Respir. Med. 2016, 4, 237–240. [Google Scholar] [CrossRef]
- Wang, C.; Tao, W.; Ni, S.; Chen, Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco. Targets. Ther. 2019, 12, 8085–8094. [Google Scholar] [CrossRef]
- Pan, Q.; Feng, Y.; Peng, Y.; Zhou, H.; Deng, Z.; Li, L.; Han, H.; Lin, J.; Shi, L.; Wang, S.; et al. Basophil Recruitment to Skin Lesions of Patients with Systemic Lupus Erythematosus Mediated by CCR1 and CCR2. Cell. Physiol. Biochem. 2017, 43, 832–839. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Opal, S.M.; Marshall, J.C.; Tracey, K.J. Sepsis definitions: Time for change. Lancet (Lond. UK) 2013, 381, 774–775. [Google Scholar] [CrossRef]
- Vincent, J.-L. The Clinical Challenge of Sepsis Identification and Monitoring. PLOS Med. 2016, 13, e1002022. [Google Scholar] [CrossRef]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock: 2012. Crit. Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Yu, W.; Ji, W.; Lin, Z.; Tan, S.; Duan, K.; Dong, Y.; Xu, L.; Li, N. Early versus delayed administration of norepinephrine in patients with septic shock. Crit. Care 2014, 18, 532. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, R.; Martin-Loeches, I.; Phillips, G.; Osborn, T.M.; Townsend, S.; Dellinger, R.P.; Artigas, A.; Schorr, C.; Levy, M.M. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: Results from a guideline-based performance improvement program. Crit. Care Med. 2014, 42, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-Y.; Choi, J.-H. Biomarkers of sepsis. Infect. Chemother. 2014, 46, 1–12. [Google Scholar] [CrossRef]
- Vincent, J.-L.; Beumier, M. Diagnostic and prognostic markers in sepsis. Expert Rev. Anti. Infect. Ther. 2013, 11, 265–275. [Google Scholar] [CrossRef]
- Reinhart, K.; Bauer, M.; Riedemann, N.C.; Hartog, C.S. New approaches to sepsis: Molecular diagnostics and biomarkers. Clin. Microbiol. Rev. 2012, 25, 609–634. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, C.; Zhang, X.; Chen, Y.; Zhang, H. MiR-145 negatively regulates TGFBR2 signaling responsible for sepsis-induced acute lung injury. Biomed. Pharmacother. 2019, 111, 852–858. [Google Scholar] [CrossRef]
- Geng, Y.; Jiang, J.; Wu, C. Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 2018, 11, 98. [Google Scholar] [CrossRef]
- Hoffmann, U.; Hoffmann, U.; Bertsch, T.; Hoffmann, U.; Bertsch, T.; Dvortsak, E.; Liebetrau, C.; Lang, S.; Liebe, V.; Huhle, G.; et al. Matrix-metalloproteinases and their inhibitors are elevated in severe sepsis: Prognostic value of TIMP-1 in severe sepsis. Scand. J. Infect. Dis. 2006, 38, 867–872. [Google Scholar] [CrossRef]
- Aguirre, A.; Blázquez-Prieto, J.; Amado-Rodriguez, L.; López-Alonso, I.; Batalla-Solís, E.; González-López, A.; Sánchez-Pérez, M.; Mayoral-Garcia, C.; Gutiérrez-Fernández, A.; Albaiceta, G.M. Matrix metalloproteinase-14 triggers an anti-inflammatory proteolytic cascade in endotoxemia. J. Mol. Med. 2017, 95, 487–497. [Google Scholar] [CrossRef]
- Huo, R.; Dai, M.; Fan, Y.; Zhou, J.-Z.; Li, L.; Zu, J. Predictive value of miRNA-29a and miRNA-10a-5p for 28-day mortality in patients with sepsis-induced acute kidney injury. Nan Fang Yi Ke Da Xue Xue Bao 2017, 37, 646–651. [Google Scholar] [PubMed]
- Huang, M.; Zhong, Z.; Lv, M.; Shu, J.; Tian, Q.; Chen, J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 2016, 7, 47186–47200. [Google Scholar] [CrossRef] [PubMed]
- Reithmair, M.; Buschmann, D.; Märte, M.; Kirchner, B.; Hagl, D.; Kaufmann, I.; Pfob, M.; Chouker, A.; Steinlein, O.K.; Pfaffl, M.W.; et al. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J. Cell. Mol. Med. 2017, 21, 2403–2411. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.-L.; McGinley, J.P.; Drysdale, S.B.; Pollard, A.J. Epidemiology and Immune Pathogenesis of Viral Sepsis. Front. Immunol. 2018, 9, 2147. [Google Scholar] [CrossRef]


| CircRNA | Mechanism | Role | Reference |
|---|---|---|---|
| mcircRASGEF1B | Inducible with LPS stimuli during microbial infection through TLR4 | Protects cells against microbial infection by regulating the stability of mature ICAM-1 mRNAs | [138] |
| Circ-010231 | Regulates different immune responses to virus | Plays an important role in host defense to virus by inducing competitive binding between host circRNAs and viral mRNAs | [126] |
| Circ_0005105 | Interacts with the mRNA of pro-inflammatory cytokines | Induces a pro-inflammatory phenotype | [103] |
| Circ RNA-CER | Induced in chondrocytes by IL-1 and TNFα | Mediates an inflammatory response through interaction with IL-1 and TNFα | [136] |
| Circ_0028644 | Regulates the expression of NF-κB | Modulates different pro-inflammatory phenotypes | [101] |
| Circ_4099 | Modulates the expression of miR-616-5p | Closely related to inflammatory phenotypes through TNF- α | [100] |
| Circ_0003159 | Regulates miR-223 | Induces an inflammatory response due to increased expression of IL-6, IL-1β, and TNF-α | [111] |
| Circ_RSF1 | Regulates the expression of inflammatory cytokines | Represses the interactions of miR-146a with RAC1 by eliminating its inhibitory effect on the RAC1 pathway | [115] |
| Circ_102685 | Modulates the expression of miR-146a | Plays a role in endotoxin tolerance, immunosuppression, inflammatory response, and antiviral pathways | [116] |
| Circ_0005075 | Regulates miR-23a-5p and miR-23b-5p | Suppresses the expression of miR-23b-5p in cancer and is related to immune response | [123,124,125] |
| Circ-PVT1 | Interacts with the miR-125 family | Exhibits a possible correlation with sepsis severity, inflammation, and increased mortality | [117] |
| Circ-GLI2 | Negatively regulates the expression of miR-125b-5p | Involved in inflammation and immune response pathways | [119] |
| Circ-MYLK and Circ_CTDP1 | Regulates miR-29a-3p | Feasible predictive biomarker for assessing 28-day mortality of sepsis patients | [139,140] |
| Circ_HIAT1 | Regulates miR-29a-3p and miR-29c-3p, and matrix metrix metalloproteinases MMP-9 and MMP-2 | Increases miRNAs stability | [141] |
| Circ_NSD2 | Regulates different processes through sponge miR-199b | Related to the low miR-199b-5p levels found in sepsis patients | [142] |
| Circ_0005785 | Regulates miR-181a and miR-181b | Possible role in sepsis by promoting immunosuppression in late sepsis | [127] |
| Circ_0000096 | Regulates the expression of miR-224 and miR-200a | Modulates the immune response through cyclin D1, CDK6, MMP-2, and MMP-9 | [143] |
| Circ_001569 | Modulates the expression of miR-145 | Involved in the immune response of host to pathogens | [139,140] |
| Circ_HIPK3 | Modulates the expression of miR-193a-3 and miR-124 | Mediates a pro-inflammatory state by modeling the inflammatory response through sponge miR-124 (inhibitor of IL-6) | [144] |
| Circ_0003528, Circ_0007196 and Circ_0078738 | Interacts with miR-192-5p | Related to the low levels found in sepsis patients | [145] |
| Circ RNA-9119 | Modulates miR-26a | Increases the expression of PTGS2 by modulating the response of endothelium | [146] |
| Circ_TRIM33 and Circ_FOXO3 | Modulates the expression of miR-191 and induces the expression of TET1 | Induces proliferation, migration, and immune regulation | [147,148,149] |
| Circ RNA_007893 | Regulates the expression of IL-6 | Regulates the expression of IL-6, through sponging and endogenous miR-485-5p | [102] |
| Circ MAN2B2 | Regulates the expression of S1000A8 | Modulates immunosuppressive states | [128] |
| Circ RNA-MSR | Modulates miR-27 | Induces pro-inflammatory phenotype | [121] |
| Circ_0012919 | Increases the expression of DNMT1 | Modulates immune response by reducing the expression of CD70 and CD11a in CD4+ T cells | [104,150] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beltrán-García, J.; Osca-Verdegal, R.; Nacher-Sendra, E.; Pallardó, F.V.; García-Giménez, J.L. Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance. Cells 2020, 9, 1544. https://doi.org/10.3390/cells9061544
Beltrán-García J, Osca-Verdegal R, Nacher-Sendra E, Pallardó FV, García-Giménez JL. Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance. Cells. 2020; 9(6):1544. https://doi.org/10.3390/cells9061544
Chicago/Turabian StyleBeltrán-García, Jesús, Rebeca Osca-Verdegal, Elena Nacher-Sendra, Federico V. Pallardó, and José Luis García-Giménez. 2020. "Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance" Cells 9, no. 6: 1544. https://doi.org/10.3390/cells9061544
APA StyleBeltrán-García, J., Osca-Verdegal, R., Nacher-Sendra, E., Pallardó, F. V., & García-Giménez, J. L. (2020). Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance. Cells, 9(6), 1544. https://doi.org/10.3390/cells9061544





