Radiation Response of Murine Embryonic Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Treatment Modalities
2.3. Colony Forming Ability
2.4. MTT Test
2.5. Cell Cycle Analysis
2.6. Immunofluorescence Staining of γH2AX
2.7. Differentiation of mESCs
2.8. RNA Isolation and Microarray Preparation
2.9. Gene Expression Analysis
2.10. Statistics
3. Results
3.1. Survival and Viability of mESCs after X-rays Exposure
3.2. Cell Cycle Progression after X-ray Exposure
3.3. γH2AX as Marker of the Cellular Response to DNA Double Strand Breaks
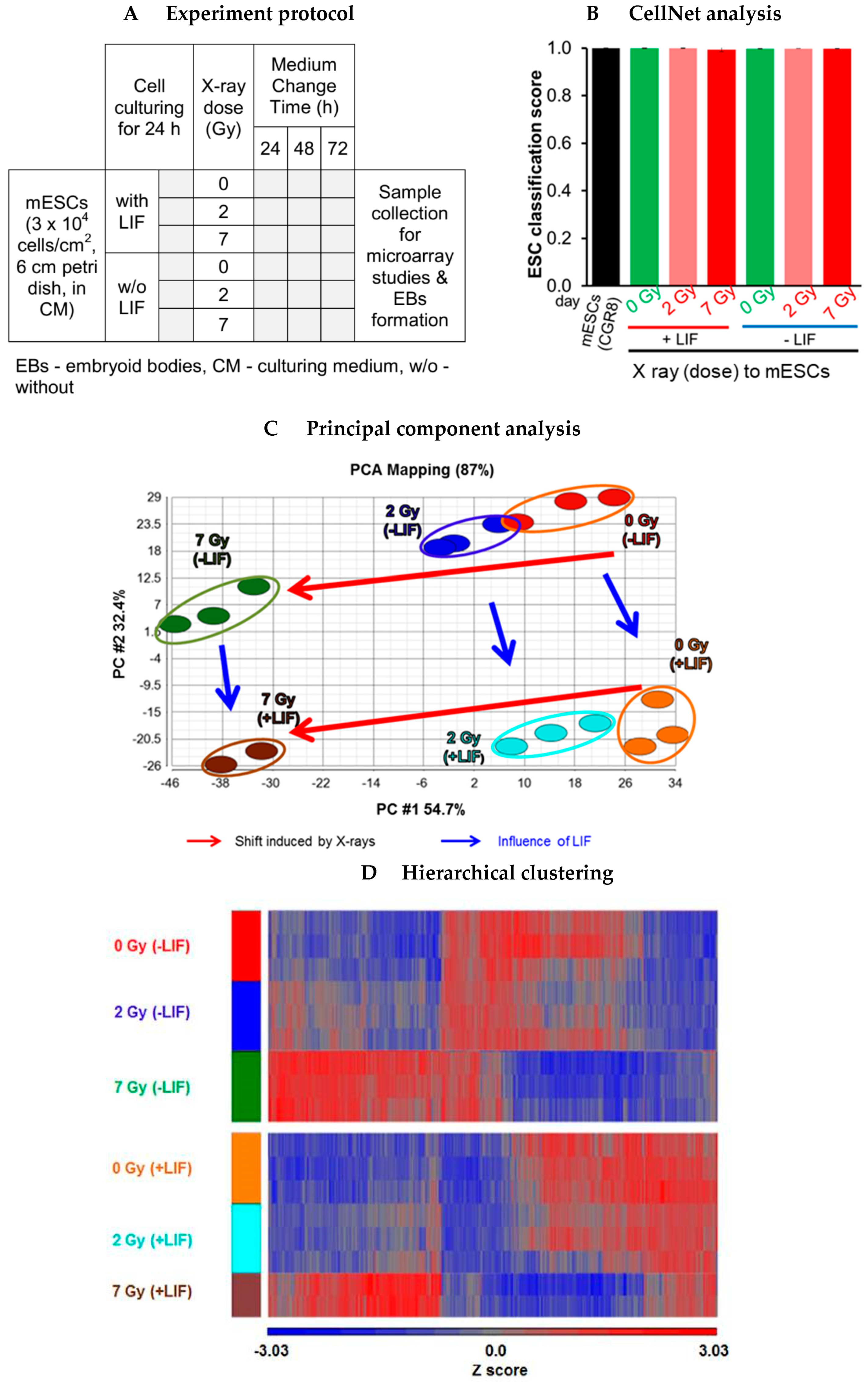
3.4. Data Structure of X-ray Deregulated Genes in mESCs Cultured in Presence and Absence of LIF
3.4.1. Microscopic Observations
3.4.2. CellNet Analysis
3.4.3. Overview of Differentially Regulated Genes
3.4.4. Developmental Genes and X-ray Influenced Genes
3.4.5. Overlap Analysis of Developmental Genes with X-ray Influenced Genes and Biological Processes Influenced (GOs)
3.4.6. Overlap Analysis of Biological Process Gene Ontologies (GOs) Influenced by Deregulated Genes
3.4.7. The Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways and Chromosome Location of Developmental Genes and X-ray Influenced Genes
3.5. Fate of X-ray Exposed mESCs after Differentiation towards Beating Cardiomyocytes
4. Discussion
4.1. Clonogenic Survival and Viability
4.2. Cell Cycle Progression and DNA Repair
4.3. Differentiation and Gene Expression Changes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Keller, G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005, 19, 1129–1155. [Google Scholar] [CrossRef] [PubMed]
- De Santis, M.; Cesari, E.; Nobili, E.; Straface, G.; Cavaliere, A.F.; Caruso, A. Radiation effects on development. Birth Defects Res. Part C Embryo Today Rev. 2007, 81, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Panyutin, I.V.; Holar, S.A.; Neumann, R.D.; Panyutin, I.G. Effect of ionizing radiation on the proliferation of human embryonic stem cells. Sci. Rep. 2017, 7, 43995. [Google Scholar] [CrossRef]
- Denissova, N.G.; Tereshchenko, I.V.; Cui, E.; Stambrook, P.J.; Shao, C.; Tischfield, J.A. Ionizing radiation is a potent inducer of mitotic recombination in mouse embryonic stem cells. Mutat. Res. 2011, 715, 1–6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Chuykin, I.A.; Lianguzova, M.S.; Pospelova, T.V.; Pospelov, V.A. Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle (Georget. Tex.) 2008, 7, 2922–2928. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A.; Chen, D.J. Ionizing radiation-induced mutation of human cells with different DNA repair capacities. Adv. Space Res. 1996, 18, 119–126. [Google Scholar] [CrossRef]
- Thomas, J.W.; LaMantia, C.; Magnuson, T. X-ray-induced mutations in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 1998, 95, 1114–1119. [Google Scholar] [CrossRef]
- Tou, J.; Ronca, A.; Grindeland, R.; Wade, C. Models to study gravitational biology of Mammalian reproduction. Biol. Reprod. 2002, 67, 1681–1687. [Google Scholar] [CrossRef]
- Serova, L.V.; Denisova, L.A. The effect of weightlessness on the reproductive function of mammals. Physiologist 1982, 25, S9–S12. [Google Scholar]
- Kojima, Y.; Sasaki, S.; Kubota, Y.; Ikeuchi, T.; Hayashi, Y.; Kohri, K. Effects of simulated microgravity on mammalian fertilization and preimplantation embryonic development in vitro. Fertil. Steril. 2000, 74, 1142–1147. [Google Scholar] [CrossRef]
- Crawford-Young, S.J. Effects of microgravity on cell cytoskeleton and embryogenesis. Int. J. Dev. Biol. 2006, 50, 183–191. [Google Scholar] [CrossRef]
- Wakayama, S.; Kawahara, Y.; Li, C.; Yamagata, K.; Yuge, L.; Wakayama, T. Detrimental effects of microgravity on mouse preimplantation development in vitro. PLoS ONE 2009, 4, e6753. [Google Scholar] [CrossRef] [PubMed]
- Wakayama, S.; Kamada, Y.; Yamanaka, K.; Kohda, T.; Suzuki, H.; Shimazu, T.; Tada, M.N.; Osada, I.; Nagamatsu, A.; Kamimura, S.; et al. Healthy offspring from freeze-dried mouse spermatozoa held on the International Space Station for 9 months. Proc. Natl. Acad. Sci. USA 2017, 114, 5988–5993. [Google Scholar] [CrossRef] [PubMed]
- Serova, L.V.; Denisova, L.A.; Apanasenko, Z.I.; Kuznetsova, M.A.; Meizerov, E.S. Reproductive function of the male rat after a flight on the Kosmos-1129 biosatellite. Kosm. Biol. I Aviakosmicheskaia Med. 1982, 16, 62–65. [Google Scholar]
- Luft, S.; Arrizabalaga, O.; Kulish, I.; Nasonova, E.; Durante, M.; Ritter, S.; Schroeder, I.S. Ionizing Radiation Alters Human Embryonic Stem Cell Properties and Differentiation Capacity by Diminishing the Expression of Activin Receptors. Stem Cells Dev. 2017, 26, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Shinde, V.; Brungs, S.; Hescheler, J.; Hemmersbach, R.; Sachinidis, A. Pipette-based Method to Study Embryoid Body Formation Derived from Mouse and Human Pluripotent Stem Cells Partially Recapitulating Early Embryonic Development Under Simulated Microgravity Conditions. Microgravity Sci. Technol. 2016, 28, 287–295. [Google Scholar] [CrossRef]
- Shinde, V.; Brungs, S.; Henry, M.; Wegener, L.; Nemade, H.; Rotshteyn, T.; Acharya, A.; Baumstark-Khan, C.; Hellweg, C.E.; Hescheler, J.; et al. Simulated Microgravity Modulates Differentiation Processes of Embryonic Stem Cells. Cell. Physiol. Biochem. 2016, 38, 1483–1499. [Google Scholar] [CrossRef]
- Acharya, A.; Brungs, S.; Henry, M.; Rotshteyn, T.; Singh Yaduvanshi, N.; Wegener, L.; Jentzsch, S.; Hescheler, J.; Hemmersbach, R.; Boeuf, H.; et al. Modulation of Differentiation Processes in Murine Embryonic Stem Cells Exposed to Parabolic Flight-Induced Acute Hypergravity and Microgravity. Stem Cells Dev 2018, 27, 838–847. [Google Scholar] [CrossRef]
- Suhring, R.; Diamond, M.L.; Scheringer, M.; Wong, F.; Pucko, M.; Stern, G.; Burt, A.; Hung, H.; Fellin, P.; Li, H.; et al. Organophosphate Esters in Canadian Arctic Air: Occurrence, Levels and Trends. Environ. Sci. Technol. 2016, 50, 7409–7415. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, V.; Horrillo, A.; Hmadcha, A.; Suarez-Alvarez, B.; Fernandez, A.F.; Lara, E.; Casado, S.; Menendez, P.; Bueno, C.; Garcia-Castro, J.; et al. Cancer genes hypermethylated in human embryonic stem cells. PLoS ONE 2008, 3, e3294. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, J.C.; Lee, J.Y.; Park, M.J. Oct4 suppresses IR-induced premature senescence in breast cancer cells through STAT3- and NF-κB-mediated IL-24 production. Int. J. Oncol. 2018, 53, 47–58. [Google Scholar] [CrossRef]
- De Waard, H.; Sonneveld, E.; de Wit, J.; Esveldt-van Lange, R.; Hoeijmakers, J.H.; Vrieling, H.; van der Horst, G.T. Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts. DNA Repair 2008, 7, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Giglia-Mari, G.; Zotter, A.; Vermeulen, W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011, 3, a000745. [Google Scholar] [CrossRef]
- Kapinas, K.; Grandy, R.; Ghule, P.; Medina, R.; Becker, K.; Pardee, A.; Zaidi, S.K.; Lian, J.; Stein, J.; van Wijnen, A.; et al. The abbreviated pluripotent cell cycle. J. Cell. Physiol. 2013, 228, 9–20. [Google Scholar] [CrossRef]
- Jacobs, K.M.; Misri, S.; Meyer, B.; Raj, S.; Zobel, C.L.; Sleckman, B.P.; Hallahan, D.E.; Sharma, G.G. Unique epigenetic influence of H2AX phosphorylation and H3K56 acetylation on normal stem cell radioresponses. Mol. Biol. Cell 2016, 27, 1332–1345. [Google Scholar] [CrossRef]
- Tichy, E.D.; Pillai, R.; Deng, L.; Liang, L.; Tischfield, J.; Schwemberger, S.J.; Babcock, G.F.; Stambrook, P.J. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010, 19, 1699–1711. [Google Scholar] [CrossRef]
- Abulizi, P.; Loganathan, N.; Zhao, D.; Mele, T.; Zhang, Y.; Zwiep, T.; Liu, K.; Zheng, X. Growth Differentiation Factor-15 Deficiency Augments Inflammatory Response and Exacerbates Septic Heart and Renal Injury Induced by Lipopolysaccharide. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Cahan, P.; Li, H.; Morris, S.A.; da Rocha, E.L.; Daley, G.Q.; Collins, J.J. CellNet: Network biology applied to stem cell engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef]
- Kim, K.P.; Scholer, H.R. CellNet—Where your cells are standing. Cell 2014, 158, 699–701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morris, S.A.; Cahan, P.; Li, H.; Zhao, A.M.; San Roman, A.K.; Shivdasani, R.A.; Collins, J.J.; Daley, G.Q. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 2014, 158, 889–902. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Hu, Y.; Hellweg, C.E.; Baumstark-Khan, C.; Reitz, G.; Lau, P. Cell cycle delay in murine pre-osteoblasts is more pronounced after exposure to high-LET compared to low-LET radiation. Radiat. Environ. Biophys. 2014, 53, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.A.; Jones, G.D.; Anderson, R.; Iyer, P.; Narayanan, D.; Sandhu, J.; Singh, R.; Talbot, C.J.; Tufarelli, C. DNMTs are required for delayed genome instability caused by radiation. Epigenetics 2012, 7, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Sioftanos, G.; Ismail, A.; Fohse, L.; Shanley, S.; Worku, M.; Short, S.C. BRCA1 and BRCA2 heterozygosity in embryonic stem cells reduces radiation-induced Rad51 focus formation but is not associated with radiosensitivity. Int. J. Radiat. Biol. 2010, 86, 1095–1105. [Google Scholar] [CrossRef]
- Banuelos, C.A.; Banath, J.P.; MacPhail, S.H.; Zhao, J.; Eaves, C.A.; O’Connor, M.D.; Lansdorp, P.M.; Olive, P.L. Mouse but not human embryonic stem cells are deficient in rejoining of ionizing radiation-induced DNA double-strand breaks. DNA Repair 2008, 7, 1471–1483. [Google Scholar] [CrossRef]
- Meyer, B.; Fabbrizi, M.R.; Raj, S.; Zobel, C.L.; Hallahan, D.E.; Sharma, G.G. Histone H3 Lysine 9 Acetylation Obstructs ATM Activation and Promotes Ionizing Radiation Sensitivity in Normal Stem Cells. Stem Cell Rep. 2016, 7, 1013–1022. [Google Scholar] [CrossRef]
- Shirai, H.; Fujimori, H.; Gunji, A.; Maeda, D.; Hirai, T.; Poetsch, A.R.; Harada, H.; Yoshida, T.; Sasai, K.; Okayasu, R.; et al. Parg deficiency confers radio-sensitization through enhanced cell death in mouse ES cells exposed to various forms of ionizing radiation. Biochem. Biophys. Res. Commun. 2013, 435, 100–106. [Google Scholar] [CrossRef]
- Jiang, J.; Belikova, N.A.; Hoye, A.T.; Zhao, Q.; Epperly, M.W.; Greenberger, J.S.; Wipf, P.; Kagan, V.E. A mitochondria-targeted nitroxide/hemigramicidin S conjugate protects mouse embryonic cells against gamma irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 816–825. [Google Scholar] [CrossRef]
- Rijkers, T.; Van Den Ouweland, J.; Morolli, B.; Rolink, A.G.; Baarends, W.M.; Van Sloun, P.P.; Lohman, P.H.; Pastink, A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998, 18, 6423–6429. [Google Scholar] [CrossRef] [PubMed]
- Shibahara, K.; Uchiumi, T.; Fukuda, T.; Kura, S.; Tominaga, Y.; Maehara, Y.; Kohno, K.; Nakabeppu, Y.; Tsuzuki, T.; Kuwano, M. Targeted disruption of one allele of the Y-box binding protein-1 (YB-1) gene in mouse embryonic stem cells and increased sensitivity to cisplatin and mitomycin C. Cancer Sci. 2004, 95, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Bassing, C.H.; Chua, K.F.; Sekiguchi, J.; Suh, H.; Whitlow, S.R.; Fleming, J.C.; Monroe, B.C.; Ciccone, D.N.; Yan, C.; Vlasakova, K.; et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 2002, 99, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Budzowska, M.; Jaspers, I.; Essers, J.; de Waard, H.; van Drunen, E.; Hanada, K.; Beverloo, B.; Hendriks, R.W.; de Klein, A.; Kanaar, R.; et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004, 23, 3548–3558. [Google Scholar] [CrossRef]
- Seifert, B.A.; Dejosez, M.; Zwaka, T.P. Ronin influences the DNA damage response in pluripotent stem cells. Stem Cell Res. 2017, 23, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, M.V.; Panyutin, I.V.; Onyshchenko, M.I.; Panyutin, I.G.; Neumann, R.D. Expression of pluripotency-associated genes in the surviving fraction of cultured human embryonic stem cells is not significantly affected by ionizing radiation. Gene 2010, 455, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; An, K.; Cai, J.; Li, G.; Yang, C.; Liu, H.; Du, F.; Han, X.; Zhang, Z.; et al. Lower genomic stability of induced pluripotent stem cells reflects increased non-homologous end joining. Cancer Commun. (Lond. Engl.) 2018, 38, 49. [Google Scholar] [CrossRef]
- Rebuzzini, P.; Pignalosa, D.; Mazzini, G.; Di Liberto, R.; Coppola, A.; Terranova, N.; Magni, P.; Redi, C.A.; Zuccotti, M.; Garagna, S. Mouse embryonic stem cells that survive gamma-rays exposure maintain pluripotent differentiation potential and genome stability. J. Cell. Physiol. 2012, 227, 1242–1249. [Google Scholar] [CrossRef]
- Suchorska, W.M.; Augustyniak, E.; Łukjanow, M. Comparison of the early response of human embryonic stem cells and human induced pluripotent stem cells to ionizing radiation. Mol. Med. Rep. 2017, 15, 1952–1962. [Google Scholar] [CrossRef]
- Momcilović, O.; Choi, S.; Varum, S.; Bakkenist, C.; Schatten, G.; Navara, C. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells (Dayt. Ohio) 2009, 27, 1822–1835. [Google Scholar] [CrossRef]
- Filion, T.M.; Qiao, M.; Ghule, P.N.; Mandeville, M.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Altieri, D.C.; Stein, G.S. Survival responses of human embryonic stem cells to DNA damage. J. Cell. Physiol. 2009, 220, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, M.V.; Panyutin, I.V.; Panyutin, I.G.; Neumann, R.D. Dynamics of the transcriptome response of cultured human embryonic stem cells to ionizing radiation exposure. Mutat. Res. 2011, 709–710, 40–48. [Google Scholar] [CrossRef]
- Stambrook, P.J.; Tichy, E.D. Preservation of genomic integrity in mouse embryonic stem cells. Adv. Exp. Med. Biol. 2010, 695, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Cervantes, R.B.; Tichy, E.; Tischfield, J.A.; Stambrook, P.J. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutat. Res. 2007, 614, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Aprelikova, O.; Pace, A.J.; Fang, B.; Koller, B.H.; Liu, E.T. BRCA1 is a selective co-activator of 14-3-3 sigma gene transcription in mouse embryonic stem cells. J. Biol. Chem. 2001, 276, 25647–25650. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Hu, Z.; An, L.; He, Y.; Hang, H. Targeted deletion of mouse Rad1 leads to deficient cellular DNA damage responses. Protein Cell 2011, 2, 410–422. [Google Scholar] [CrossRef][Green Version]
- Dolezalova, D.; Mraz, M.; Barta, T.; Plevova, K.; Vinarsky, V.; Holubcova, Z.; Jaros, J.; Dvorak, P.; Pospisilova, S.; Hampl, A. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells (Dayt. Ohio) 2012, 30, 1362–1372. [Google Scholar] [CrossRef]
- Van der Laan, S.; Tsanov, N.; Crozet, C.; Maiorano, D. High Dub3 expression in mouse ESCs couples the G1/S checkpoint to pluripotency. Mol. Cell 2013, 52, 366–379. [Google Scholar] [CrossRef]
- Koledova, Z.; Kafkova, L.R.; Kramer, A.; Divoky, V. DNA damage-induced degradation of Cdc25A does not lead to inhibition of Cdk2 activity in mouse embryonic stem cells. Stem Cells (Dayt. Ohio) 2010, 28, 450–461. [Google Scholar] [CrossRef]
- Hong, Y.; Stambrook, P.J. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc. Natl. Acad. Sci. USA 2004, 101, 14443–14448. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L. Detection of DNA damage in individual cells by analysis of histone H2AX phosphorylation. Methods Cell Biol. 2004, 75, 355–373. [Google Scholar] [CrossRef] [PubMed]
- Maynard, S.; Swistowska, A.M.; Lee, J.W.; Liu, Y.; Liu, S.T.; Da Cruz, A.B.; Rao, M.; de Souza-Pinto, N.C.; Zeng, X.; Bohr, V.A. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells (Dayt. Ohio) 2008, 26, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.R.; Golding, S.E.; Rao, R.R.; Valerie, K. Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS ONE 2010, 5, e10001. [Google Scholar] [CrossRef]
- Serrano, L.; Liang, L.; Chang, Y.; Deng, L.; Maulion, C.; Nguyen, S.; Tischfield, J.A. Homologous recombination conserves DNA sequence integrity throughout the cell cycle in embryonic stem cells. Stem Cells Dev. 2011, 20, 363–374. [Google Scholar] [CrossRef]
- Hennicke, T.; Nieweg, K.; Brockmann, N.; Kassack, M.U.; Gottmann, K.; Fritz, G. mESC-based in vitro differentiation models to study vascular response and functionality following genotoxic insults. Toxicol. Sci. 2015, 144, 138–150. [Google Scholar] [CrossRef]
- Jacquet, P.; de Saint-Georges, L.; Vankerkom, J.; Baugnet-Mahieu, L. Embryonic death, dwarfism and fetal malformations after irradiation of embryos at the zygote stage: Studies on two mouse strains. Mutat. Res. 1995, 332, 73–87. [Google Scholar] [CrossRef]
- Luft, S.; Pignalosa, D.; Nasonova, E.; Arrizabalaga, O.; Helm, A.; Durante, M.; Ritter, S. Fate of D3 mouse embryonic stem cells exposed to X-rays or carbon ions. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 760, 56–63. [Google Scholar] [CrossRef]
- Sokolov, M.V.; Neumann, R.D. Radiation-induced bystander effects in cultured human stem cells. PLoS ONE 2010, 5, e14195. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Yoshida, K.; Kitada, K.; Kizu, A.; Tachibana, D.; Fukui, M.; Morita, T.; Koyama, M. Low-dose irradiation of mouse embryos increases Smad-p21 pathway activity and preserves pluripotency. J. Assist. Reprod. Genet. 2018, 35, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.D.; Sun, N.; Huang, M.; Zhang, W.Y.; Lee, A.S.; Li, Z.; Wang, S.X.; Wu, J.C. Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res. 2010, 70, 5539–5548. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, M.R.; Knudsen, T.B.; Kellems, R.E. Genetically engineered mice demonstrate that adenosine deaminase is essential for early postimplantation development. Development (Camb. Engl.) 1997, 124, 3089–3097. [Google Scholar]
- De Luca, A.; De Falco, M.; De Luca, L.; Penta, R.; Shridhar, V.; Baldi, F.; Campioni, M.; Paggi, M.G.; Baldi, A. Pattern of expression of HtrA1 during mouse development. J. Histochem. Cytochem. 2004, 52, 1609–1617. [Google Scholar] [CrossRef]
- Zaghlool, A.; Halvardson, J.; Zhao, J.J.; Etemadikhah, M.; Kalushkova, A.; Konska, K.; Jernberg-Wiklund, H.; Thuresson, A.C.; Feuk, L. A Role for the Chromatin-Remodeling Factor BAZ1A in Neurodevelopment. Hum. Mutat. 2016, 37, 964–975. [Google Scholar] [CrossRef]
- Jacquier, A.; Delorme, C.; Belotti, E.; Juntas-Morales, R.; Solé, G.; Dubourg, O.; Giroux, M.; Maurage, C.A.; Castellani, V.; Rebelo, A.; et al. Cryptic amyloidogenic elements in mutant NEFH causing Charcot-Marie-Tooth 2 trigger aggresome formation and neuronal death. Acta Neuropathol. Commun. 2017, 5, 1–15. [Google Scholar] [CrossRef]
- Côté, F.; Collard, J.F.; Julien, J.P. Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: A mouse model of amyotrophic lateral sclerosis. Cell 1993, 73, 35–46. [Google Scholar] [CrossRef]
- Gil-Cayuela, C.; López, A.; Martínez-Dolz, L.; González-Juanatey, J.R.; Lago, F.; Roselló-Lletí, E.; Rivera, M.; Portolés, M. The altered expression of autophagy-related genes participates in heart failure: NRBP2 and CALCOCO2 are associated with left ventricular dysfunction parameters in human dilated cardiomyopathy. PLoS ONE 2019, 14, e0215818. [Google Scholar] [CrossRef]
- Dekaban, A.S. Effects of x-radiation on mouse fetus during gesttion: Emphasis on distribution of cerebral lesions, Part II. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 1969, 10, 68–77. [Google Scholar]
- Verreet, T.; Quintens, R.; Van Dam, D.; Verslegers, M.; Tanori, M.; Casciati, A.; Neefs, M.; Leysen, L.; Michaux, A.; Janssen, A.; et al. A multidisciplinary approach unravels early and persistent effects of X-ray exposure at the onset of prenatal neurogenesis. J. Neurodev. Disord. 2015, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Solozobova, V.; Rolletschek, A.; Blattner, C. Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol. 2009, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, Y.; Dubois, W.; Wu, X.; Shi, J.; Huang, J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell 2012, 46, 30–42. [Google Scholar] [CrossRef] [PubMed]











| Group | KEGG Pathway (Up) | Total Genes | p Value | KEGG Pathway (Down) | Total Genes | p Value |
|---|---|---|---|---|---|---|
| w/o LIF 7 Gy vs. 0 Gy | p53 signaling | 7 | 7.74 × 10−4 | Glycolysis | 8 | 3.62 × 10−6 |
| Hippo signaling | 11 | 1.80 × 10−4 | Pyruvate metabolism | 6 | 3.82 × 10−5 | |
| PI3K-Akt signaling | 15 | 1.81 × 10−3 | Biosynthesis of amino acids | 7 | 1.17 × 10−4 | |
| with LIF 7 Gy vs. 0 Gy | p53 signaling | 8 | 2.13 × 10−4 | Glycolysis | 8 | 4.31 × 10−5 |
| Pyruvate metabolism | 5 | 2.34 × 10−3 | ||||
| Biosynthesis of amino acids | 8 | 1.27 × 10−4 | ||||
| w/o LIF 0 Gy vs. with LIF 0 Gy | Axon guidance | 5 | 1.97 × 10−2 | Pluripotency signaling | 7 | 3.17 × 10−3 |
| Osteoclast differentiation | 6 | 1.02 × 10−2 |
| Group | Gene Regulation | Total Genes | Chromosome Number |
|---|---|---|---|
| w/o LIF 7 Gy vs. 0 Gy | Up | 32 | 3 |
| with LIF 7 Gy vs. 0 Gy | Up | 43 | 3 |
| Up | 42 | 13 | |
| w/o LIF 0 Gy vs. with LIF 0 Gy | Up | 13 | 15 |
| Down | 21 | 11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hellweg, C.E.; Shinde, V.; Srinivasan, S.P.; Henry, M.; Rotshteyn, T.; Baumstark-Khan, C.; Schmitz, C.; Feles, S.; Spitta, L.F.; Hemmersbach, R.; et al. Radiation Response of Murine Embryonic Stem Cells. Cells 2020, 9, 1650. https://doi.org/10.3390/cells9071650
Hellweg CE, Shinde V, Srinivasan SP, Henry M, Rotshteyn T, Baumstark-Khan C, Schmitz C, Feles S, Spitta LF, Hemmersbach R, et al. Radiation Response of Murine Embryonic Stem Cells. Cells. 2020; 9(7):1650. https://doi.org/10.3390/cells9071650
Chicago/Turabian StyleHellweg, Christine E., Vaibhav Shinde, Sureshkumar Perumal Srinivasan, Margit Henry, Tamara Rotshteyn, Christa Baumstark-Khan, Claudia Schmitz, Sebastian Feles, Luis F. Spitta, Ruth Hemmersbach, and et al. 2020. "Radiation Response of Murine Embryonic Stem Cells" Cells 9, no. 7: 1650. https://doi.org/10.3390/cells9071650
APA StyleHellweg, C. E., Shinde, V., Srinivasan, S. P., Henry, M., Rotshteyn, T., Baumstark-Khan, C., Schmitz, C., Feles, S., Spitta, L. F., Hemmersbach, R., Hescheler, J., & Sachinidis, A. (2020). Radiation Response of Murine Embryonic Stem Cells. Cells, 9(7), 1650. https://doi.org/10.3390/cells9071650







