BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. sgRNA Selection, sgRNA and Cas9 mRNA Synthesis
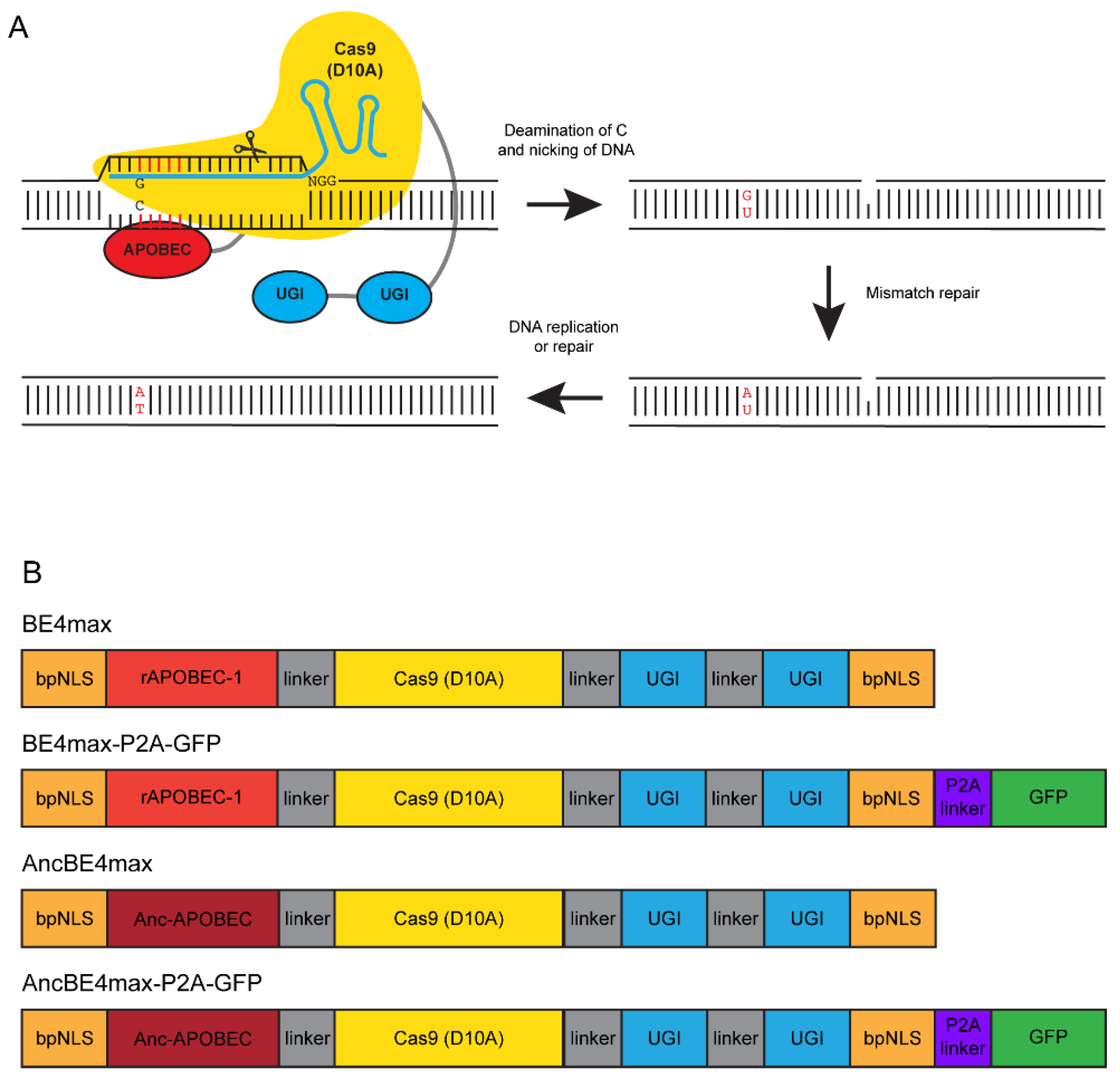
2.3. Base Editing Constructs and RNA Synthesis
2.4. Zebrafish Injections
2.5. DNA Extraction, Flourescent PCR for CRISPR-STAT and Sanger Sequencing
2.6. Somatic Base Editing Analysis
2.7. Screening for Germline Transmission of Edited Nucleotides
3. Results
3.1. Experimental Design
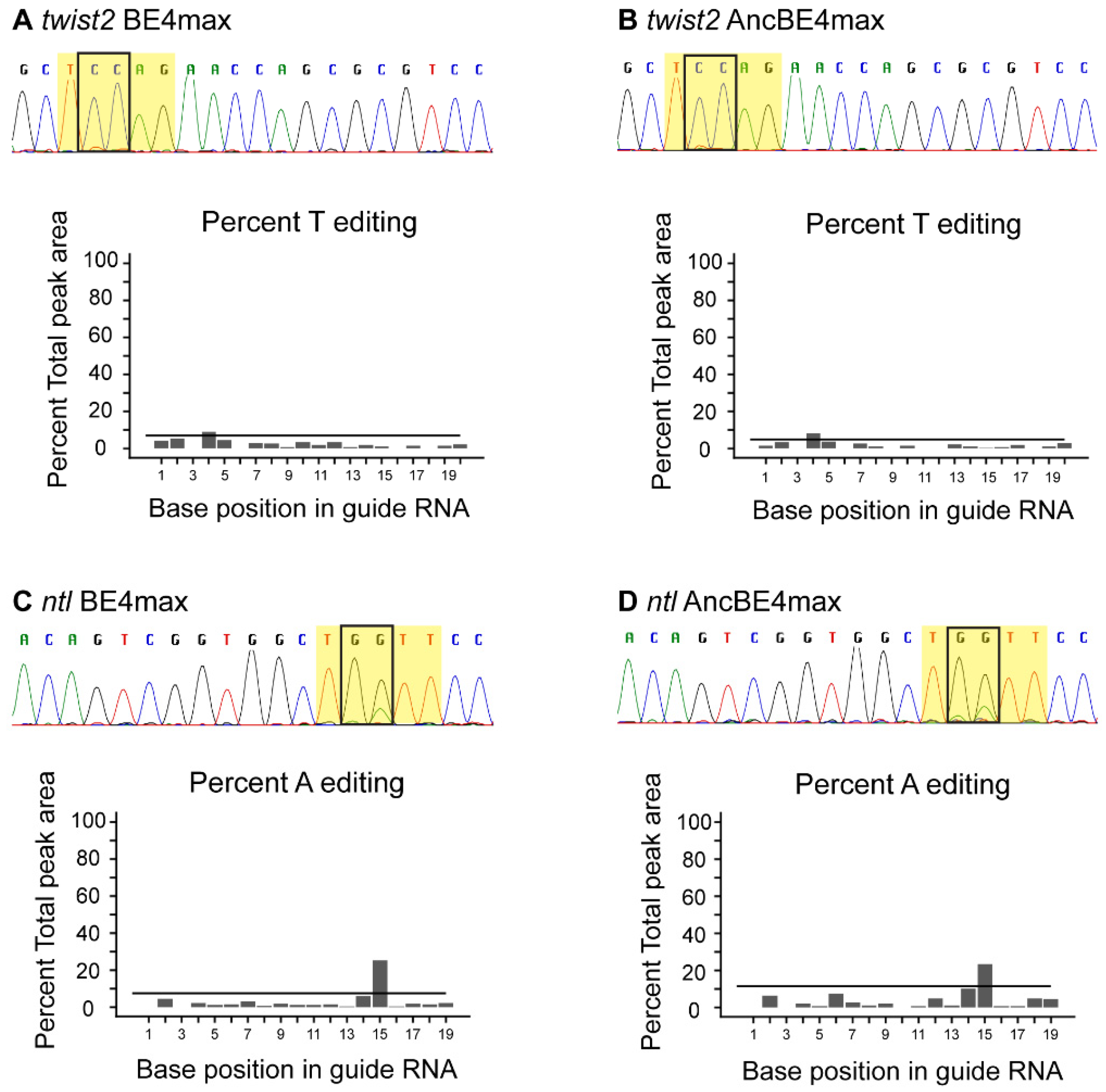
3.2. Somatic Base Editing
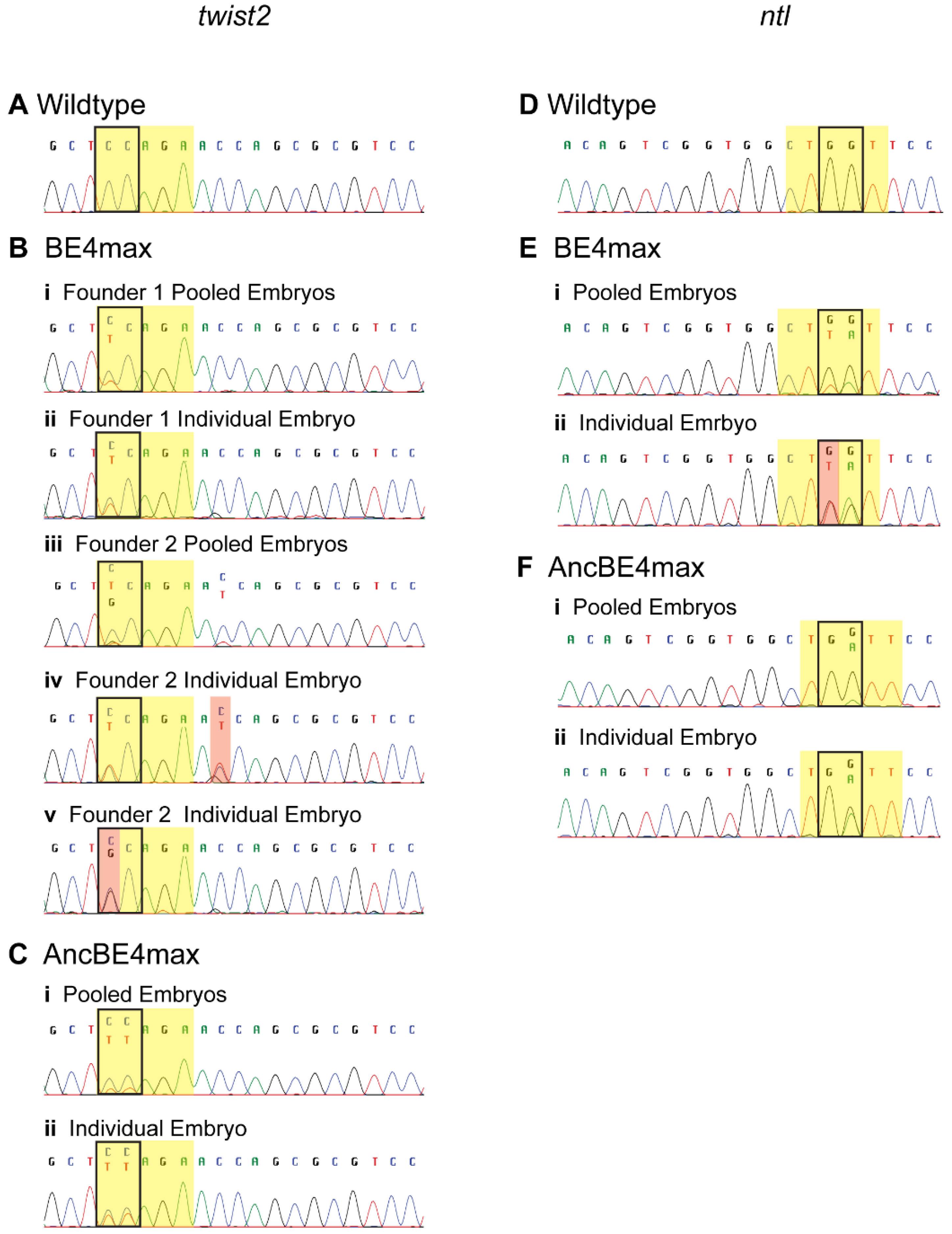
3.3. Germline Transmission of Edited Nucleotides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Petree, C.; Requena, T.; Varshney, P.; Varshney, G.K. Expanding the crispr toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol. 2019, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubbini, D.; Cornet, C.; Terriente, J.; Di Donato, V. Crispr meets zebrafish: Accelerating the discovery of new therapeutic targets. SLAS Discov. 2020, 2472555220926920. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, L.; Page-McCaw, P.S.; Chen, W. Zebrafish genome engineering using the crispr-cas9 system. Trends Genet. 2016, 32, 815–827. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Akhmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient mutagenesis by cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide rnas. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using crispr/cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [Green Version]
- Vejnar, C.E.; Moreno-Mateos, M.A.; Cifuentes, D.; Bazzini, A.A.; Giraldez, A.J. Optimized crispr-cas9 system for genome editing in zebrafish. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Unal Eroglu, A.; Mulligan, T.S.; Zhang, L.; White, D.T.; Sengupta, S.; Nie, C.; Lu, N.Y.; Qian, J.; Xu, L.; Pei, W.; et al. Multiplexed crispr/cas9 targeting of genes implicated in retinal regeneration and degeneration. Front. Cell Dev. Biol. 2018, 6, 88. [Google Scholar] [CrossRef] [Green Version]
- DiNapoli, S.E.; Martinez-McFaline, R.; Gribbin, C.K.; Wrighton, P.J.; Balgobin, C.A.; Nelson, I.; Leonard, A.; Maskin, C.R.; Shwartz, A.; Quenzer, E.D.; et al. Synthetic crispr/cas9 reagents facilitate genome editing and homology directed repair. Nucleic Acids Res. 2020, 48, e38. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Quintans, B.; Ordonez-Ugalde, A.; Cacheiro, P.; Carracedo, A.; Sobrido, M.J. Medical genomics: The intricate path from genetic variant identification to clinical interpretation. Appl. Transl. Genom. 2014, 3, 60–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilbeck, K.; Quinlan, A.; Yandell, M. Settling the score: Variant prioritization and mendelian disease. Nat. Rev. Genet. 2017, 18, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Scott, A.J.; Brandt, M.; Hall, I.M. Genomic analysis in the age of human genome sequencing. Cell 2019, 177, 70–84. [Google Scholar] [CrossRef] [Green Version]
- Anna, A.; Monika, G. Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2018, 59, 253–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, G.A.; Liao, M.; You, Z.; Lissouba, A.; Chen, B.E.; Drapeau, P. Homology directed knockin of point mutations in the zebrafish tardbp and fus genes in als using the crispr/cas9 system. PLoS ONE 2016, 11, e0150188. [Google Scholar] [CrossRef]
- Prykhozhij, S.V.; Fuller, C.; Steele, S.L.; Veinotte, C.J.; Razaghi, B.; Robitaille, J.M.; McMaster, C.R.; Shlien, A.; Malkin, D.; Berman, J.N. Optimized knock-in of point mutations in zebrafish using crispr/cas9. Nucleic Acids Res. 2018, 46, e102. [Google Scholar] [CrossRef]
- Tessadori, F.; Roessler, H.I.; Savelberg, S.M.C.; Chocron, S.; Kamel, S.M.; Duran, K.J.; van Haelst, M.M.; van Haaften, G.; Bakkers, J. Effective crispr/cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, Z.; Ge, W. An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by crispr/cas9-mediated gene editing. J. Biol. Chem. 2018, 293, 6611–6622. [Google Scholar] [CrossRef] [Green Version]
- Albadri, S.; Del Bene, F.; Revenu, C. Genome editing using crispr/cas9-based knock-in approaches in zebrafish. Methods 2017, 121–122, 77–85. [Google Scholar] [CrossRef]
- Boel, A.; De Saffel, H.; Steyaert, W.; Callewaert, B.; De Paepe, A.; Coucke, P.J.; Willaert, A. Crispr/cas9-mediated homology-directed repair by ssodns in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prykhozhij, S.V.; Caceres, L.; Berman, J.N. New developments in crispr/cas-based functional genomics and their implications for research using zebrafish. Curr. Gene Ther. 2017, 17, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Komor, A.C.; Yeh, W.H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 15790. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Qin, W.; Lu, X.; Xu, J.; Huang, H.; Bai, H.; Li, S.; Lin, S. Programmable base editing of zebrafish genome using a modified crispr-cas9 system. Nat. Commun. 2017, 8, 118. [Google Scholar] [CrossRef]
- Qin, W.; Lu, X.; Lin, S. Programmable base editing in zebrafish using a modified crispr-cas9 system. Methods 2018, 150, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Lu, X.; Liu, Y.; Bai, H.; Li, S.; Lin, S. Precise a*t to g*c base editing in the zebrafish genome. BMC Biol. 2018, 16, 139. [Google Scholar] [CrossRef] [PubMed]
- Simone, B.W.; Martinez-Galvez, G.; WareJoncas, Z.; Ekker, S.C. Fishing for understanding: Unlocking the zebrafish gene editor’s toolbox. Methods 2018, 150, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Yoshioka, S.; Nishida, K.; Hosokawa, H.; Kakizuka, A.; Maegawa, S. In vivo targeted single-nucleotide editing in zebrafish. Sci Rep. 2018, 8, 11423. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with crispr-cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of a*t to g*c in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, J.; Yin, W.; Zhang, Z.; Song, Y.; Chang, X. Targeted aid-mediated mutagenesis (tam) enables efficient genomic diversification in mammalian cells. Nat. Methods 2016, 13, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Komor, A.C.; Levy, J.M.; Packer, M.S.; Zhao, K.T.; Liu, D.R. Increasing the genome-targeting scope and precision of base editing with engineered cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017, 35, 371–376. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved cas9 variants with broad pam compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Liu, Y.; Yang, B.; Wang, X.; Wei, J.; Lu, Z.; Zhang, Y.; Wu, J.; Huang, X.; et al. Base editing with a cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. [Google Scholar] [CrossRef]
- Zafra, M.P.; Schatoff, E.M.; Katti, A.; Foronda, M.; Breinig, M.; Schweitzer, A.Y.; Simon, A.; Han, T.; Goswami, S.; Montgomery, E.; et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018, 36, 888–893. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a crispr-cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. Crispr/cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage mu gam protein yields c:G-to-t:A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerfield, M. The Zebrafish Book, a Guide for the Laboratory Use of Zebrafish (Danio Rerio), 5th ed.; M. Westerfield: Eugene, OR, USA, 2007. [Google Scholar]
- Varshney, G.K.; Carrington, B.; Pei, W.; Bishop, K.; Chen, Z.; Fan, C.; Xu, L.; Jones, M.; LaFave, M.C.; Ledin, J.; et al. A high-throughput functional genomics workflow based on crispr/cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 2016, 11, 2357–2375. [Google Scholar] [CrossRef] [PubMed]
- Carrington, B.; Varshney, G.K.; Burgess, S.M.; Sood, R. Crispr-stat: An easy and reliable pcr-based method to evaluate target-specific sgrna activity. Nucleic Acids Res. 2015, 43, e157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Kaini, P.; Sander, J.D.; Joung, J.K.; Peterson, R.T.; Yeh, J.R. Heritable and precise zebrafish genome editing using a crispr-cas system. PLoS ONE 2013, 8, e68708. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.E.; Frangakis, S.; Katsanis, N. Interpreting human genetic variation with in vivo zebrafish assays. Biochim. Biophys. Acta 2014, 1842, 1960–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, G.K.; Sood, R.; Burgess, S.M. Understanding and editing the zebrafish genome. Adv. Genet. 2015, 92, 1–52. [Google Scholar]
- Gehrke, J.M.; Cervantes, O.; Clement, M.K.; Wu, Y.; Zeng, J.; Bauer, D.E.; Pinello, L.; Joung, J.K. An apobec3a-cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018, 36, 977–982. [Google Scholar] [CrossRef]
- Lee, H.K.; Smith, H.E.; Liu, C.; Willi, M.; Hennighausen, L. Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun. Biol. 2020, 3, 19. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Lim, K.; Kim, S.T.; Yoon, S.H.; Kim, K.; Ryu, S.M.; Kim, J.S. Genome-wide target specificities of crispr rna-guided programmable deaminases. Nat. Biotechnol. 2017, 35, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Sun, Y.; Wei, W.; Yuan, T.; Ying, W.; Sun, H.; Yuan, L.; Steinmetz, L.M.; Li, Y.; Yang, H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019, 364, 289–292. [Google Scholar] [CrossRef]
- Doman, J.L.; Raguram, A.; Newby, G.A.; Liu, D.R. Evaluation and minimization of cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020, 38, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a crispr nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billon, P.; Bryant, E.E.; Joseph, S.A.; Nambiar, T.S.; Hayward, S.B.; Rothstein, R.; Ciccia, A. Crispr-mediated base editing enables efficient disruption of eukaryotic genes through induction of stop codons. Mol. Cell 2017, 67, 1068–1079 e1064. [Google Scholar] [CrossRef] [Green Version]
- Kuscu, C.; Parlak, M.; Tufan, T.; Yang, J.; Szlachta, K.; Wei, X.; Mammadov, R.; Adli, M. Crispr-stop: Gene silencing through base-editing-induced nonsense mutations. Nat. Methods 2017, 14, 710–712. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.; Mulligan, T.S.; Shen, M.C.; Wang, H.; Scahill, C.M.; Tan, F.J.; Du, S.J.; Busch-Nentwich, E.M.; Farber, S.A. Mrna processing in mutant zebrafish lines generated by chemical and crispr-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 2017, 13, e1007105. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, D. Fish mutant, where is thy phenotype? PLoS Genet. 2018, 14, e1007197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gapinske, M.; Luu, A.; Winter, J.; Woods, W.S.; Kostan, K.A.; Shiva, N.; Song, J.S.; Perez-Pinera, P. Crispr-skip: Programmable gene splicing with single base editors. Genome Biol. 2018, 19, 107. [Google Scholar] [CrossRef] [Green Version]

| Gene-Target | Sequence (5′-3′) |
|---|---|
| twist2-T1 | GCTC4C5AGAACCAGCGCGTCC TGG |
| twist2-T2 | GCCGC5TC7GCGTACGTTCGCC AGG |
| ntl (tbxta)1 | GGAAC5C6AGCCACCGACTGT TGG |
| Target | Base Editor | Embryos Screened | Embryos with Nucleotide Changes in Target Window | |
|---|---|---|---|---|
| Correct | Incorrect | |||
| twist2 | BE4max | 47 | 2 (4%) | None |
| ntl | 47 | 2 (4%) | None | |
| twist2 | AncBE4max | 47 | 5 (11%) | None |
| ntl | 47 | 3 (6%) | None | |
| Target | Base Editor | F0 Fish Screened 1 | F0 Fish with Base Editing | Individual F1 Embryos | |
|---|---|---|---|---|---|
| Correct Editing | Incorrect Editing | ||||
| twist2 | BE4max | 7 | 2 | 3/34 (9%) | None |
| 1/37 (3%) 2 | 8 (22%) | ||||
| AncBE4max | 4 | 1 | 7/82 (9%) | 3 (4%) | |
| ntl | BE4max | 6 | 1 | 5/122 (4%) 3 | 5 (4%) 3 |
| AncBE4max | 4 | 1 | 7/95 (7%) | None | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrington, B.; Weinstein, R.N.; Sood, R. BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish. Cells 2020, 9, 1690. https://doi.org/10.3390/cells9071690
Carrington B, Weinstein RN, Sood R. BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish. Cells. 2020; 9(7):1690. https://doi.org/10.3390/cells9071690
Chicago/Turabian StyleCarrington, Blake, Rachel N. Weinstein, and Raman Sood. 2020. "BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish" Cells 9, no. 7: 1690. https://doi.org/10.3390/cells9071690
APA StyleCarrington, B., Weinstein, R. N., & Sood, R. (2020). BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish. Cells, 9(7), 1690. https://doi.org/10.3390/cells9071690





