Fission Yeast Polarization: Modeling Cdc42 Oscillations, Symmetry Breaking, and Zones of Activation and Inhibition
Abstract
:1. Introduction
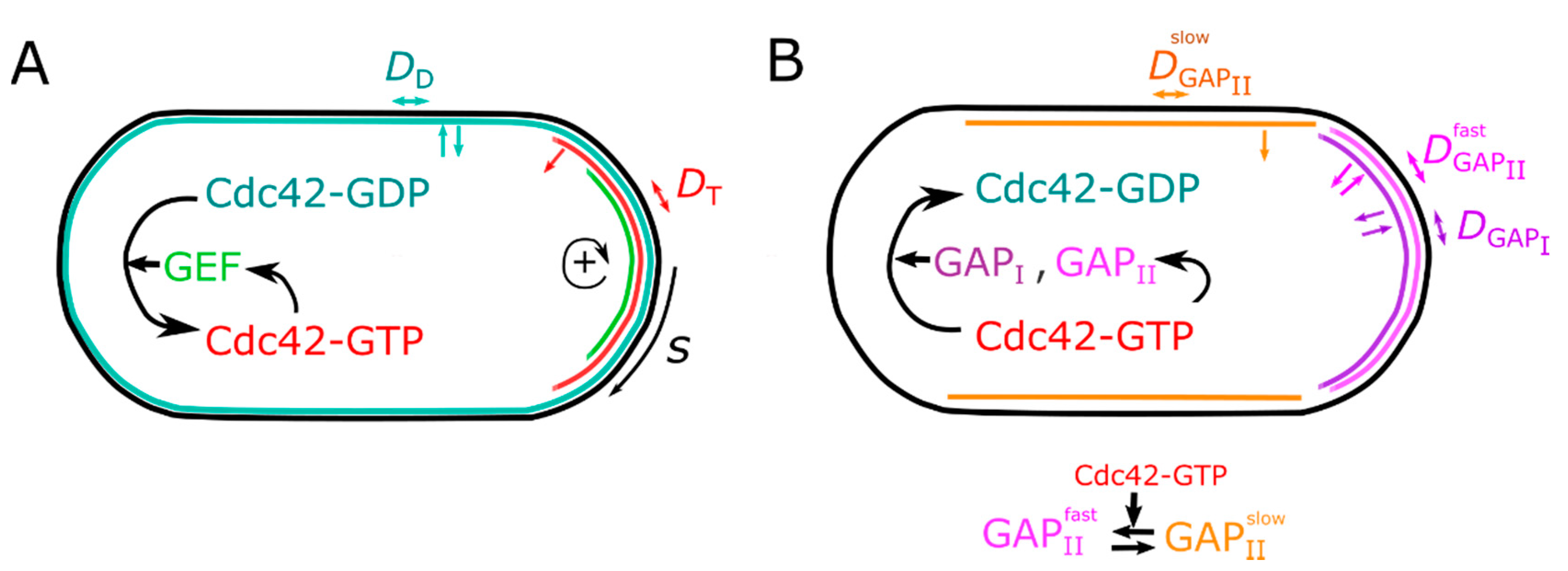
2. Model
3. Results
3.1. Goals
3.2. Dynamical States Observed in Parameter Scan
3.3. Structure of Solutions in Parameter Space
3.4. Simulations with Reduced GAPII Recruitment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Devreotes, P.N.; Bhattacharya, S.; Edwards, M.; Iglesias, P.A.; Lampert, T.; Miao, Y. Excitable Signal Transduction Networks in Directed Cell Migration. Annu. Rev. Cell Dev. Biol. 2017, 33, 103–125. [Google Scholar] [CrossRef]
- Pichaud, F.; Walther, R.F.; Nunes de Almeida, F. Regulation of Cdc42 and its effectors in epithelial morphogenesis. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [Green Version]
- Holcomb, P.S.; Deerinck, T.J.; Ellisman, M.H.; Spirou, G.A. Construction of a polarized neuron. J. Physiol. 2013, 591, 3145–3150. [Google Scholar] [CrossRef]
- Martin, S.G.; Arkowitz, R.A. Cell polarization in budding and fission yeasts. FEMS Microbiol. Rev. 2014, 38, 228–253. [Google Scholar] [CrossRef] [Green Version]
- Goryachev, A.B.; Leda, M. Many roads to symmetry breaking: molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell 2017, 28, 370–380. [Google Scholar] [CrossRef]
- Halatek, J.; Brauns, F.; Frey, E. Self-organization principles of intracellular pattern formation. Philos. Trans. R. Soc. Lond B. Biol. Sci. 2018, 373. [Google Scholar] [CrossRef]
- Pruyne, D.; Bretscher, A. Polarization of cell growth in yeast. J. Cell Sci. 2000, 113, 571–585. [Google Scholar]
- Bi, E.; Park, H.O. Cell polarization and cytokinesis in budding yeast. Genetics 2012, 191, 347–387. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.G. Spontaneous cell polarization: Feedback control of Cdc42 GTPase breaks cellular symmetry. Bioessays 2015, 37, 1193–1201. [Google Scholar] [CrossRef] [Green Version]
- Chiou, J.G.; Balasubramanian, M.K.; Lew, D.J. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 2017, 33, 77–101. [Google Scholar] [CrossRef]
- Goryachev, A.B.; Pokhilko, A.V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008, 582, 1437–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jilkine, A.; Edelstein-Keshet, L. A Comparison of Mathematical Models for Polarization of Single Eukaryotic Cells in Response to Guided Cues. PLoS Comput. Biol. 2010, 7, e1001121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein-Keshet, L.; Holmes, W.R.; Zajac, M.; Dutot, M. From simple to detailed models for cell polarization. Philos. Trans. R. Soc. Lond B. Biol. Sci. 2013, 368, 20130003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savage, N.S.; Layton, A.T.; Lew, D.J. Mechanistic mathematical model of polarity in yeast. Mol. Biol. Cell 2012, 23, 1998–2013. [Google Scholar] [CrossRef] [PubMed]
- Dyer, J.M.; Savage, N.S.; Jin, M.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 2013, 23, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Freisinger, T.; Klunder, B.; Johnson, J.; Muller, N.; Pichler, G.; Beck, G.; Costanzo, M.; Boone, C.; Cerione, R.A.; Frey, E.; et al. Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat. Commun. 2013, 4, 1807. [Google Scholar] [CrossRef] [Green Version]
- Klunder, B.; Freisinger, T.; Wedlich-Soldner, R.; Frey, E. GDI-mediated cell polarization in yeast provides precise spatial and temporal control of Cdc42 signaling. PLoS Comput. Biol. 2013, 9, e1003396. [Google Scholar] [CrossRef] [Green Version]
- Woods, B.; Lai, H.; Wu, C.F.; Zyla, T.R.; Savage, N.S.; Lew, D.J. Parallel Actin-Independent Recycling Pathways Polarize Cdc42 in Budding Yeast. Curr. Biol. 2016, 26, 2114–2126. [Google Scholar] [CrossRef] [Green Version]
- Verde, F.; Mata, J.; Nurse, P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 1995, 131, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Mitchison, J.M.; Nurse, P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1985, 75, 357–376. [Google Scholar]
- Csikász-Nagy, A.; Gyorffy, B.; Alt, W.; Tyson, J.J.; Novák, B. Spatial controls for growth zone formation during the fission yeast cell cycle. Yeast 2008, 25, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Drake, T.; Wiley, D.J.; Buchwald, P.; Vavylonis, D.; Verde, F. Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science 2012, 337, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerone, L.; Novak, B.; Neufeld, Z. Mathematical model for growth regulation of fission yeast Schizosaccharomyces pombe. PLoS ONE 2012, 7, e49675. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Jilkine, A. Modeling the Dynamics of Cdc42 Oscillation in Fission Yeast. Biophys. J. 2018, 114, 2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, M.; Verde, F. Role of Cdc42 dynamics in the control of fission yeast cell polarization. Biochem. Soc. Trans. 2013, 41, 1745–1749. [Google Scholar] [CrossRef] [Green Version]
- Raskin, D.M.; de Boer, P.A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 4971–4976. [Google Scholar] [CrossRef] [Green Version]
- Loose, M.; Kruse, K.; Schwille, P. Protein self-organization: lessons from the min system. Annu. Rev. Biophys. 2011, 40, 315–336. [Google Scholar] [CrossRef]
- Bendezu, F.O.; Martin, S.G. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 2013, 23, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Haupt, A.; Ershov, D.; Minc, N. A Positive Feedback between Growth and Polarity Provides Directional Persistency and Flexibility to the Process of Tip Growth. Curr. Biol. 2018, 28, 3342–3351. [Google Scholar] [CrossRef] [Green Version]
- Rincon, S.; Coll, P.M.; Perez, P. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle 2007, 6, 1687–1691. [Google Scholar] [CrossRef]
- Das, M.; Wiley, D.J.; Chen, X.; Shah, K.; Verde, F. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 2009, 19, 1314–1319. [Google Scholar] [CrossRef] [Green Version]
- Kelly, F.D.; Nurse, P. Spatial control of Cdc42 activation determines cell width in fission yeast. Mol. Biol. Cell 2011, 22, 3801–3811. [Google Scholar] [CrossRef]
- Bendezu, F.O.; Vincenzetti, V.; Vavylonis, D.; Wyss, R.; Vogel, H.; Martin, S.G. Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 2015, 13, e1002097. [Google Scholar] [CrossRef] [Green Version]
- Hercyk, B.S.; Rich-Robinson, J.; Mitoubsi, A.S.; Harrell, M.A.; Das, M.E. A novel interplay between GEFs orchestrates Cdc42 activity during cell polarity and cytokinesis in fission yeast. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Lamas, I.; Merlini, L.; Vjestica, A.; Vincenzetti, V.; Martin, S.G. Optogenetics reveals Cdc42 local activation by scaffold-mediated positive feedback and Ras GTPase. PLoS Biol. 2020, 18, e3000600. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Wiley, D.J.; Medina, S.; Vincent, H.A.; Larrea, M.; Oriolo, A.; Verde, F. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell 2007, 18, 2090–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, H.; Bidone, T.C.; Vavylonis, D. Computational model of polarized actin cables and cytokinetic actin ring formation in budding yeast. Cytoskeleton (Hoboken) 2015, 72, 517–533. [Google Scholar] [CrossRef] [Green Version]
- Tatebe, H.; Nakano, K.; Maximo, R.; Shiozaki, K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 2008, 18, 322–330. [Google Scholar] [CrossRef] [Green Version]
- Revilla-Guarinos, M.T.; Martin-Garcia, R.; Villar-Tajadura, M.A.; Estravis, M.; Coll, P.M.; Perez, P. Rga6 is a Fission Yeast Rho GAP Involved in Cdc42 Regulation of Polarized Growth. Mol. Biol. Cell 2016, 27, 1524–1535. [Google Scholar] [CrossRef] [Green Version]
- Gallo Castro, D.; Martin, S.G. Differential GAP requirement for Cdc42-GTP polarization during proliferation and sexual reproduction. J. Cell Biol. 2018, 217, 4215–4229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalili, B.; Merlini, L.; Vincenzetti, V.; Martin, S.G.; Vavylonis, D. Exploration and stabilization of Ras1 mating zone: A mechanism with positive and negative feedbacks. PLoS Comput. Biol. 2018, 14, e1006317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.; Bressloff, P.C. A PDE-DDE Model for Cell Polarization in Fission Yeast. SIAM J. Appl. Math. 2016, 76, 1844–1870. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Kang, H.W.; Jilkine, A. Comparison of Deterministic and Stochastic Regime in a Model for Cdc42 Oscillations in Fission Yeast. Bull. Math. Biol. 2019, 81, 1268–1302. [Google Scholar] [CrossRef] [PubMed]
- Paquin-Lefebvre, F.; Xu, B.; DiPietro, K.L.; Lindsay, A.E.; Jilkine, A. Pattern formation in a coupled membrane-bulk reaction-diffusion model for intracellular polarization and oscillations. J. Theor. Biol. 2020, 497, 110242. [Google Scholar] [CrossRef] [Green Version]
- Geymonat, M.; Chessel, A.; Dodgson, J.; Punter, H.; Horns, F.; Nagy, A.C.; Carazo Salas, R.E. Activation of polarized cell growth by inhibition of cell polarity. bioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Merlini, L.; Khalili, B.; Dudin, O.; Michon, L.; Vincenzetti, V.; Martin, S.G. Inhibition of Ras activity coordinates cell fusion with cell-cell contact during yeast mating. J. Cell Biol. 2018, 217, 1467–1483. [Google Scholar] [CrossRef] [Green Version]
- Novak, I.L.; Gao, F.; Choi, Y.S.; Resasco, D.; Schaff, J.C.; Slepchenko, B.M. Diffusion on a Curved Surface Coupled to Diffusion in the Volume: Application to Cell Biology. J. Comput. Phys. 2007, 226, 1271–1290. [Google Scholar] [CrossRef] [Green Version]
- Kokkoris, K.; Gallo Castro, D.; Martin, S.G. The Tea4-PP1 landmark promotes local growth by dual Cdc42 GEF recruitment and GAP exclusion. J. Cell Sci. 2014, 127, 2005–2016. [Google Scholar] [CrossRef] [Green Version]
- Ng, A.Y.E.; Ng, A.Q.E.; Zhang, D. ER-PM Contacts Restrict Exocytic Sites for Polarized Morphogenesis. Curr. Biol. 2018, 28, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Tay, Y.D.; Leda, M.; Goryachev, A.B.; Sawin, K.E. Local and global Cdc42 guanine nucleotide exchange factors for fission yeast cell polarity are coordinated by microtubules and the Tea1-Tea4-Pom1 axis. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [Green Version]
- Howell, A.S.; Jin, M.; Wu, C.F.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 2012, 149, 322–333. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Nunez, I.; Rodriguez, M.; Wiley, D.J.; Rodriguez, J.; Sarkeshik, A.; Yates, J.R., 3rd; Buchwald, P.; Verde, F. Phosphorylation-dependent inhibition of Cdc42 GEF Gef1 by 14-3-3 protein Rad24 spatially regulates Cdc42 GTPase activity and oscillatory dynamics during cell morphogenesis. Mol. Biol. Cell 2015, 26, 3520–3534. [Google Scholar] [CrossRef] [PubMed]
- Pablo, M.; Ramirez, S.A.; Elston, T.C. Particle-based simulations of polarity establishment reveal stochastic promotion of Turing pattern formation. PLoS Comput. Biol. 2018, 14, e1006016. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.R.; Nuñez, I.; Chen, C.; Das, M.E.; Wiley, D.J.; D’Urso, G.; Buchwald, P.; Vavylonis, D.; Verde, F. Cdc42 GTPase Activating Proteins (GAPs) Maintain Generational Inheritance of Cell Polarity and Cell Shape in Fission Yeast. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kuo, C.C.; Savage, N.S.; Chen, H.; Wu, C.F.; Zyla, T.R.; Lew, D.J. Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr. Biol. 2014, 24, 753–759. [Google Scholar] [CrossRef] [Green Version]
- Rapali, P.; Mitteau, R.; Braun, C.; Massoni-Laporte, A.; Unlu, C.; Bataille, L.; Arramon, F.S.; Gygi, S.P.; McCusker, D. Scaffold-mediated gating of Cdc42 signalling flux. Elife 2017, 6. [Google Scholar] [CrossRef]
- Chen, C.; Rodriguez Pino, M.; Haller, P.R.; Verde, F. Conserved NDR/LATS kinase controls RAS GTPase activity to regulate cell growth and chronological lifespan. Mol. Biol. Cell 2019, 30, 2598–2616. [Google Scholar] [CrossRef]
- Tay, Y.D.; Leda, M.; Spanos, C.; Rappsilber, J.; Goryachev, A.B.; Sawin, K.E. Fission Yeast NDR/LATS Kinase Orb6 Regulates Exocytosis via Phosphorylation of the Exocyst Complex. Cell Rep. 2019, 26, 1654–1667. [Google Scholar] [CrossRef] [Green Version]
- Okada, S.; Leda, M.; Hanna, J.; Savage, N.S.; Bi, E.; Goryachev, A.B. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell 2013, 26, 148–161. [Google Scholar] [CrossRef] [Green Version]
- Drake, T.; Vavylonis, D. Model of fission yeast cell shape driven by membrane-bound growth factors and the cytoskeleton. PLoS Comput. Biol. 2013, 9, e1003287. [Google Scholar] [CrossRef] [Green Version]
- Bement, W.M.; Leda, M.; Moe, A.M.; Kita, A.M.; Larson, M.E.; Golding, A.E.; Pfeuti, C.; Su, K.C.; Miller, A.L.; Goryachev, A.B.; et al. Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat. Cell Biol. 2015, 17, 1471–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandin, G.; Marenduzzo, D.; Goryachev, A.B.; Orlandini, E. Curvature-driven positioning of Turing patterns in phase-separating curved membranes. Soft Matter 2016, 12, 3888–3896. [Google Scholar] [CrossRef] [PubMed]
- Brauns, F.; Pawlik, G.; Halatek, J.; Kerssemakers, J.; Frey, E.; Dekker, C. Bulk-surface coupling reconciles Min-protein pattern formation in vitro and in vivo. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]







| Variable | Reference Value | Description |
|---|---|---|
| 0.02 μm2/s | Diffusion coefficient of Cdc42-GTP from [33] | |
| 0.2 μm2/s | Diffusion coefficient of Cdc42-GDP from [33] | |
| 0.03 μm2/s | Diffusion coefficient of GAPI, estimated | |
| 0.0625 μm2/s | Diffusion coefficient of fast GAPII, estimated | |
| 0.005 μm2/s | Diffusion coefficient of slow GAPII, estimated | |
| 0.000625/s | Spontaneous rate Cdc42-GTP hydrolysis, adjusted | |
| 0.00325 μm2/s | Rate constant of GAPI-mediated Cdc42-GTP hydrolysis, adjusted | |
| 0.00125 μm2/s | Rate constant of fast GAPII-mediated Cdc42-GTP hydrolysis, adjusted | |
| 250/s | Rate constant of Cdc42-GTP-mediated GAPI recruitment, adjusted | |
| 0.03/s | Rate constant of Cdc42-GTP-mediated GAPII recruitment, adjusted | |
| 2/s | Rate of fast GAPII conversion to slow form, adjusted | |
| 0.025 μm2/s | Cdc42-GTP-mediated conversion of slow to fast GAPII, adjusted | |
| 0.0005 μm2/s | Rate constant of slow GAPII-mediated Cdc42-GTP hydrolysis, adjusted | |
| 600/μm2 | Saturating concentration of GAPI negative feedback, adjusted | |
| 0.0025 μm2/s | Rate constant of GEF-mediated Cdc42-GDP activations, adjusted | |
| 0.5 μm3 | Linear rate constant of GEF recruitment to Cdc42-GTP, adjusted | |
| 0.1 μm5 | Quadratic rate constant of GEF recruitment to Cdc42-GTP, adjusted | |
| 250 | Total pool of GEFs, estimated | |
| 2.4/s/μm2 | Flux of Cdc42-GDP from cytoplasm to membrane, estimated | |
| 0.005/s | Rate of Cdc42-GTP dissociation from membrane from [33] | |
| 0.03/s | Rate of Cdc42-GDP dissociation from membrane from [33] | |
| 0.01/s | Rate of GAPI dissociation from membrane, adjusted | |
| 0.0125/s | Rate of fast GAPII dissociation from membrane, adjusted | |
| 0.0025/s | Rate of slow GAPII dissociation from membrane, adjusted | |
| 0.0021/s | Rate of random Cdc42-GDP conversion to Cdc42-GTP, adjusted | |
| 2.5 μm | GEF activation scale at cell tips, estimated | |
| L | 8 μm | Cell length |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalili, B.; Lovelace, H.D.; Rutkowski, D.M.; Holz, D.; Vavylonis, D. Fission Yeast Polarization: Modeling Cdc42 Oscillations, Symmetry Breaking, and Zones of Activation and Inhibition. Cells 2020, 9, 1769. https://doi.org/10.3390/cells9081769
Khalili B, Lovelace HD, Rutkowski DM, Holz D, Vavylonis D. Fission Yeast Polarization: Modeling Cdc42 Oscillations, Symmetry Breaking, and Zones of Activation and Inhibition. Cells. 2020; 9(8):1769. https://doi.org/10.3390/cells9081769
Chicago/Turabian StyleKhalili, Bita, Hailey D. Lovelace, David M. Rutkowski, Danielle Holz, and Dimitrios Vavylonis. 2020. "Fission Yeast Polarization: Modeling Cdc42 Oscillations, Symmetry Breaking, and Zones of Activation and Inhibition" Cells 9, no. 8: 1769. https://doi.org/10.3390/cells9081769





