A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2
Abstract
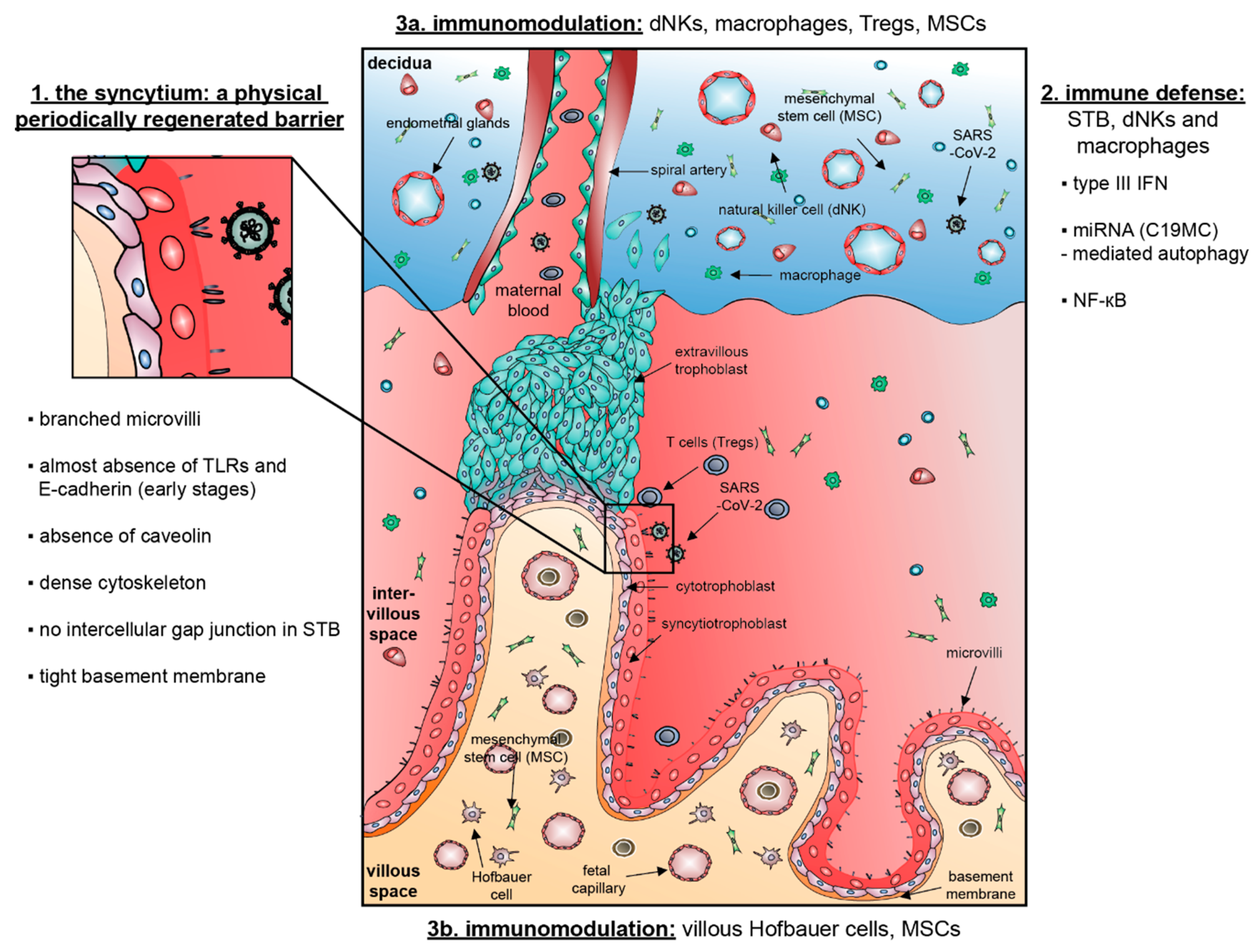
:1. Introduction
2. The Placenta in Virus Defense
2.1. A Structural and Physical Barrier for SARS-CoV-2
2.2. Pathogens May Pass through the Placental Barrier
3. SARS-CoV-2 and Pregnancy
3.1. The Placenta: Receptors and Proteases for SARS-CoV-2 Entry
3.2. Potential Evidence for Vertical Transmission of SARS-CoV-2
3.3. Placental Pathology Caused by SARS-CoV-2
4. General Immune Defense Pathways of the Human Placenta
4.1. The Placenta: Crucial Molecular Signaling Pathways against Viruses
4.1.1. The Type III IFN Signaling in Immune Defense
4.1.2. Trophoblastic microRNAs and Autophagy in Immune Defense
4.1.3. The Nuclear Factor Kappa B (NF-κB) Pathway in Immune Defense
5. Perspectives for Future Investigations
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bedford, J.; Enria, D.; Giesecke, J.; Heymann, D.L.; Ihekweazu, C.; Kobinger, G.; Lane, H.C.; Memish, Z.; Oh, M.D.; Sall, A.A.; et al. COVID-19: Towards controlling of a pandemic. Lancet 2020, 395, 1015–1018. [Google Scholar] [CrossRef]
- WHO Coronavirus Disease (COVID-19) Dashboard. World Health Organization: Geneva, Switzerland. Available online: https://covid19.who.int/ (accessed on 2 July 2020).
- Silasi, M.; Cardenas, I.; Kwon, J.Y.; Racicot, K.; Aldo, P.; Mor, G. Viral infections during pregnancy. Am. J. Reprod. Immunol. 2015, 73, 199–213. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.K.; Oh, S.J.; Park, H.; Shin, O.S. Recent Updates on Research Models and Tools to Study Virus-Host Interactions at the Placenta. Viruses 2019, 12, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouillet, J.F.; Ouyang, Y.; Bayer, A.; Coyne, C.B.; Sadovsky, Y. The role of trophoblastic microRNAs in placental viral infection. Int. J. Dev. Biol. 2014, 58, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Longman, R.E.; Johnson, T.R. Viral respiratory disease in pregnancy. Curr. Opin. Obstet. Gynecol. 2007, 19, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.S.; Zheng, B.J.; Li, Y.M.; Poon, L.L.M.; Xie, Z.H.; Chan, K.H.; Li, P.H.; Tan, S.Y.; Chang, Q.; Xie, J.P.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Channappanavar, R.; Perlman, S. Middle East Respiratory Syndrome: Emergence of a Pathogenic Human Coronavirus. Annu. Rev. Med. 2017, 68, 387–399. [Google Scholar] [CrossRef] [Green Version]
- Knight, M.; Bunch, K.; Vousden, N.; Morris, E.; Simpson, N.; Gale, C.; O’Brien, P.; Quigley, M.; Brocklehurst, P.; Kurinczuk, J.J.; et al. Characteristics and outcomes of pregnant women admitted to hospital with confirmed SARS-CoV-2 infection in UK: National population based cohort study. BMJ 2020, 369, m2107. [Google Scholar] [CrossRef]
- Lokken, E.M.; Walker, C.L.; Delaney, S.; Kachikis, A.; Kretzer, N.M.; Erickson, A.; Resnick, R.; Vanderhoeven, J.; Hwang, J.K.; Barnhart, N.; et al. Clinical characteristics of 46 pregnant women with a severe acute respiratory syndrome coronavirus 2 infection in Washington State. Am. J. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef]
- Kayem, G.; Lecarpentier, E.; Deruelle, P.; Bretelle, F.; Azria, E.; Blanc, J.; Bohec, C.; Bornes, M.; Ceccaldi, P.F.; Chalet, Y.; et al. A snapshot of the Covid-19 pandemic among pregnant women in France. J. Gynecol. Obstet. Hum. Reprod. 2020, 101826. [Google Scholar] [CrossRef]
- Sentilhes, L.; De Marcillac, F.; Jouffrieau, C.; Kuhn, P.; Thuet, V.; Hansmann, Y.; Ruch, Y.; Fafi-Kremer, S.; Deruelle, P. COVID-19 in pregnancy was associated with maternal morbidity and preterm birth. Am. J. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L. The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knofler, M.; Pollheimer, J. Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, J.R.; Bakardjiev, A.I. Pathogens and the placental fortress. Curr. Opin. Microbiol. 2012, 15, 36–43. [Google Scholar] [CrossRef] [Green Version]
- O’Tierney-Ginn, P.F.; Lash, G.E. Beyond pregnancy: Modulation of trophoblast invasion and its consequences for fetal growth and long-term children’s health. J. Reprod. Immunol. 2014, 104–105, 37–42. [Google Scholar] [CrossRef]
- Manaster, I.; Mandelboim, O. The unique properties of uterine NK cells. Am. J. Reprod. Immunol. 2010, 63, 434–444. [Google Scholar] [CrossRef]
- Ventura Ferreira, M.S.; Bienert, M.; Muller, K.; Rath, B.; Goecke, T.; Oplander, C.; Braunschweig, T.; Mela, P.; Brummendorf, T.H.; Beier, F.; et al. Comprehensive characterization of chorionic villi-derived mesenchymal stromal cells from human placenta. Stem. Cell Res. Ther. 2018, 9, 28. [Google Scholar] [CrossRef]
- Leon-Juarez, M.; Martinez-Castillo, M.; Gonzalez-Garcia, L.D.; Helguera-Repetto, A.C.; Zaga-Clavellina, V.; Garcia-Cordero, J.; Flores-Pliego, A.; Herrera-Salazar, A.; Vazquez-Martinez, E.R.; Reyes-Munoz, E. Cellular and molecular mechanisms of viral infection in the human placenta. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.; Sadovsky, Y.; Dermody, T.S.; Coyne, C.B. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017, 21, 561–567. [Google Scholar] [CrossRef]
- Malassine, A.; Frendo, J.L.; Evain-Brion, D. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 2003, 9, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Benirschke, K.; Kaufmann, P.; Baergen, R.N. Pathology of the Human Placenta, 5th ed.; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Robbins, J.R.; Skrzypczynska, K.M.; Zeldovich, V.B.; Kapidzic, M.; Bakardjiev, A.I. Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes. PLoS Pathog. 2010, 6, e1000732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ockleford, C.D.; Wakely, J.; Badley, R.A. Morphogenesis of human placental chorionic villi: Cytoskeletal, syncytioskeletal and extracellular matrix proteins. Proc. R. Soc. Lond. B Biol. Sci. 1981, 212, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Zeldovich, V.B.; Clausen, C.H.; Bradford, E.; Fletcher, D.A.; Maltepe, E.; Robbins, J.R.; Bakardjiev, A.I. Placental syncytium forms a biophysical barrier against pathogen invasion. PLoS Pathog. 2013, 9, e1003821. [Google Scholar] [CrossRef] [Green Version]
- Abrahams, V.M.; Bole-Aldo, P.; Kim, Y.M.; Straszewski-Chavez, S.L.; Chaiworapongsa, T.; Romero, R.; Mor, G. Divergent trophoblast responses to bacterial products mediated by TLRs. J. Immunol. 2004, 173, 4286–4296. [Google Scholar] [CrossRef] [Green Version]
- Holmlund, U.; Cebers, G.; Dahlfors, A.R.; Sandstedt, B.; Bremme, K.; Ekstrom, E.S.; Scheynius, A. Expression and regulation of the pattern recognition receptors Toll-like receptor-2 and Toll-like receptor-4 in the human placenta. Immunology 2002, 107, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Anderson, C.L.; Robinson, J.M. The expression of caveolin-1 and the distribution of caveolae in the murine placenta and yolk sac: Parallels to the human placenta. Placenta 2010, 31, 144–150. [Google Scholar] [CrossRef]
- Lyden, T.W.; Anderson, C.L.; Robinson, J.M. The endothelium but not the syncytiotrophoblast of human placenta expresses caveolae. Placenta 2002, 23, 640–652. [Google Scholar] [CrossRef]
- Smart, E.J.; Graf, G.A.; McNiven, M.A.; Sessa, W.C.; Engelman, J.A.; Scherer, P.E.; Okamoto, T.; Lisanti, M.P. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 1999, 19, 7289–7304. [Google Scholar] [CrossRef] [Green Version]
- Linton, E.A.; Rodriguez-Linares, B.; Rashid-Doubell, F.; Ferguson, D.J.; Redman, C.W. Caveolae and caveolin-1 in human term villous trophoblast. Placenta 2003, 24, 745–757. [Google Scholar] [CrossRef]
- Celik, O.; Saglam, A.; Baysal, B.; Derwig, I.E.; Celik, N.; Ak, M.; Aslan, S.N.; Ulas, M.; Ersahin, A.; Tayyar, A.T.; et al. Factors preventing materno-fetal transmission of SARS-CoV-2. Placenta 2020, 97, 1–5. [Google Scholar] [CrossRef]
- Lv, X.J.; Li, Y.Y.; Zhang, Y.J.; Mao, M.; Qian, G.S. Over-expression of caveolin-1 aggravate LPS-induced inflammatory response in AT-1 cells via up-regulation of cPLA2/p38 MAPK. Inflamm. Res. 2010, 59, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Aplin, J.D.; Jones, C.J.; Harris, L.K. Adhesion molecules in human trophoblast—A review. I. Villous trophoblast. Placenta 2009, 30, 293–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heerema-McKenney, A. Defense and infection of the human placenta. APMIS 2018, 126, 570–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, Y.; Boyd, C.A.; Sargent, I.L.; Redman, C.W. Hypoxia alters expression and function of syncytin and its receptor during trophoblast cell fusion of human placental BeWo cells: Implications for impaired trophoblast syncytialisation in pre-eclampsia. Biochim. Biophys. Acta 2003, 1638, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, D.A. The Origins and Emergence of Zika Virus, the Newest TORCH Infection: What’s Old Is New Again. Arch. Pathol. Lab. Med. 2017, 141, 18–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delorme-Axford, E.; Sadovsky, Y.; Coyne, C.B. The Placenta as a Barrier to Viral Infections. Annu. Rev. Virol. 2014, 1, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Schaulies, J. Cellular receptors for viruses: Links to tropism and pathogenesis. J. Gen. Virol. 2000, 81, 1413–1429. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Greber, U.F. Principles of Virus Uncoating: Cues and the Snooker Ball. Traffic 2016, 17, 569–592. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2, 109–122. [Google Scholar] [CrossRef]
- Mayor, S.; Pagano, R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 603–612. [Google Scholar] [CrossRef]
- Thorley, J.A.; McKeating, J.A.; Rappoport, J.Z. Mechanisms of viral entry: Sneaking in the front door. Protoplasma 2010, 244, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Malek, A.; Sager, R.; Kuhn, P.; Nicolaides, K.H.; Schneider, H. Evolution of maternofetal transport of immunoglobulins during human pregnancy. Am J. Reprod. Immunol. 1996, 36, 248–255. [Google Scholar] [CrossRef]
- Simoni, M.K.; Jurado, K.A.; Abrahams, V.M.; Fikrig, E.; Guller, S. Zika virus infection of Hofbauer cells. Am. J. Reprod. Immunol. 2017, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M. Apoptotic changes occur in syncytiotrophoblast of human placental villi where fibrin type fibrinoid is deposited at discontinuities in the villous trophoblast. Placenta 1996, 17, 387–391. [Google Scholar] [CrossRef]
- Sharp, A.N.; Heazell, A.E.; Crocker, I.P.; Mor, G. Placental apoptosis in health and disease. Am. J. Reprod. Immunol. 2010, 64, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Hulswit, R.J.; de Haan, C.A.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Z.; Yang, L.; Lian, X.; Xie, Y.; Li, S.; Xin, S.; Cao, P.; Lu, J. The MERS-CoV Receptor DPP4 as a Candidate Binding Target of the SARS-CoV-2 Spike. iScience 2020, 23, 101160. [Google Scholar] [CrossRef]
- Vankadari, N.; Wilce, J.A. Emerging WuHan (COVID-19) coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020, 9, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhou, Y.-S.; Lian, J.-Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.-Y.; Geng, J.-J.; et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, G.; Neves, L.A.; Anton, L.; Corthorn, J.; Chacon, C.; Germain, A.M.; Merrill, D.C.; Ferrario, C.M.; Sarao, R.; Penninger, J.; et al. Distribution of angiotensin-(1-7) and ACE2 in human placentas of normal and pathological pregnancies. Placenta 2006, 27, 200–207. [Google Scholar] [CrossRef]
- Pringle, K.G.; Tadros, M.A.; Callister, R.J.; Lumbers, E.R. The expression and localization of the human placental prorenin/renin-angiotensin system throughout pregnancy: Roles in trophoblast invasion and angiogenesis? Placenta 2011, 32, 956–962. [Google Scholar] [CrossRef]
- Li, M.; Chen, L.; Zhang, J.; Xiong, C.; Li, X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS ONE 2020, 15, e0230295. [Google Scholar] [CrossRef] [Green Version]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polanski, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Fan, X.; Wang, R.; Lu, X.; Dang, Y.L.; Wang, H.; Lin, H.Y.; Zhu, C.; Ge, H.; Cross, J.C.; et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018, 28, 819–832. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zheng, Y.; Liu, X.; Yan, L.; Fan, X.; Yong, J.; Hu, Y.; Dong, J.; Li, Q.; Wu, X.; et al. Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart. Cell Rep. 2019, 26, 1934–1950.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Ashray, N.; Bhide, A.; Chakarborty, P.; Colaco, S.; Mishra, A.; Chhabria, K.; Jolly, M.K.; Modi, D. Single-Cell RNA-seq Identifies Cell Subsets in Human Placenta That Highly Expresses Factors to Drive Pathogenesis of SARS-CoV-2. Preprints 2020, 2020050195. [Google Scholar] [CrossRef]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.A.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colaco, S.; Chhabria, K.; Singh, N.; Bhide, A.; Singh, D.; Singh, A.; Husein, A.; Mishra, A.; Sharma, R.; Ashary, N.; et al. Expression of SARS-CoV-2 receptor ACE2 and the spike protein processing enzymes in developing human embryos. arXiv 2020, arXiv:2004.04935. [Google Scholar]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef]
- Stirparo, G.G.; Boroviak, T.; Guo, G.; Nichols, J.; Smith, A.; Bertone, P. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.L.; Duan, T.; Jin, L. Single-Cell RNA Expression Profiling of ACE2 and AXL in the Human Maternal–Fetal Interface. Reprod. Dev. Med. 2020, 4, 7–10. [Google Scholar]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Luca, F.; Xu, Y.; Alazizi, A.; Leng, Y.; Hsu, C.D.; Gomez-Lopez, N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife 2020, 9. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Romero, R.; Kim, C.J.; Tarca, A.L.; Chhauy, S.; LaJeunesse, C.; Lee, D.C.; Draghici, S.; Gotsch, F.; Kusanovic, J.P.; et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: Implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. J. Immunol. 2009, 182, 3919–3927. [Google Scholar] [CrossRef] [PubMed]
- Toft, J.H.; Lian, I.A.; Tarca, A.L.; Erez, O.; Espinoza, J.; Eide, I.P.; Bjorge, L.; Draghici, S.; Romero, R.; Austgulen, R. Whole-genome microarray and targeted analysis of angiogenesis-regulating gene expression (ENG, FLT1, VEGF, PlGF) in placentas from pre-eclamptic and small-for-gestational-age pregnancies. J. Matern. Fetal Neonatal Med. 2008, 21, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Constantino, F.B.; Cury, S.S.; Nogueira, C.R.; Carvalho, R.F.; Justulin, L.A. Prediction of non-canonical routes for SARS-CoV-2 infection in human placenta cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mikheev, A.M.; Nabekura, T.; Kaddoumi, A.; Bammler, T.K.; Govindarajan, R.; Hebert, M.F.; Unadkat, J.D. Profiling gene expression in human placentae of different gestational ages: An OPRU Network and UW SCOR Study. Reprod. Sci. 2008, 15, 866–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghadhanfar, E.; Alsalem, A.; Al-Kandari, S.; Naser, J.; Babiker, F.; Al-Bader, M. The role of ACE2, angiotensin-(1-7) and Mas1 receptor axis in glucocorticoid-induced intrauterine growth restriction. Reprod. Biol. Endocrinol. 2017, 15, 97. [Google Scholar] [CrossRef] [Green Version]
- Neves, L.A.; Stovall, K.; Joyner, J.; Valdes, G.; Gallagher, P.E.; Ferrario, C.M.; Merrill, D.C.; Brosnihan, K.B. ACE2 and ANG-(1-7) in the rat uterus during early and late gestation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R151–R161. [Google Scholar] [CrossRef] [Green Version]
- Vaswani, K.; Chan, H.W.; Verma, P.; Dekker Nitert, M.; Peiris, H.N.; Wood-Bradley, R.J.; Armitage, J.A.; Rice, G.E.; Mitchell, M.D. The rat placental renin-angiotensin system—A gestational gene expression study. Reprod. Biol. Endocrinol. 2015, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, H.; Matsumoto, H.; Sato, Y.; Horie, A.; Ono, M.; Nakamura, M.; Mizumoto, Y.; Kagami, K.; Fujiwara, T.; Hattori, A.; et al. Factors Regulating Human Extravillous Trophoblast Invasion: Chemokine-peptidase and CD9-integrin Systems. Curr. Pharm. Biotechnol. 2018, 19, 764–770. [Google Scholar] [CrossRef]
- Li, K.; Nowak, R.A. The role of basigin in reproduction. Reproduction 2019. [Google Scholar] [CrossRef]
- Chen, L.; Belton, R.J., Jr.; Nowak, R.A. Basigin-mediated gene expression changes in mouse uterine stromal cells during implantation. Endocrinology 2009, 150, 966–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Alfaidy, N.; Challis, J.R. Expression of extracellular matrix metalloproteinase inducer in human placenta and fetal membranes at term labor. J. Clin. Endocrinol. Metab. 2004, 89, 2897–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, M.; Ohtani, H.; Satoh, H.; Matsuoka, S.; Hori, S.; Fujii, T.; Taketani, Y.; Sawada, Y. Characterization of transplacental transfer of paroxetine in perfused human placenta: Development of a pharmacokinetic model to evaluate tapered dosing. Drug Metab. Dispos. 2013, 41, 2124–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, Y.; Li, W.; Tran, V.; Khalil, R.A. EMMPRIN-mediated induction of uterine and vascular matrix metalloproteinases during pregnancy and in response to estrogen and progesterone. Biochem. Pharmacol. 2013, 86, 734–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.L.; Lam, M.P.; Lam, K.K.; Leung, C.O.; Pang, R.T.; Chu, I.K.; Wan, T.H.; Chai, J.; Yeung, W.S.; Chiu, P.C. Identification of CD147 (basigin) as a mediator of trophoblast functions. Hum. Reprod. 2013, 28, 2920–2929. [Google Scholar] [CrossRef] [Green Version]
- Amati, E.; Perbellini, O.; Rotta, G.; Bernardi, M.; Chieregato, K.; Sella, S.; Rodeghiero, F.; Ruggeri, M.; Astori, G. High-throughput immunophenotypic characterization of bone marrow- and cord blood-derived mesenchymal stromal cells reveals common and differentially expressed markers: Identification of angiotensin-converting enzyme (CD143) as a marker differentially expressed between adult and perinatal tissue sources. Stem. Cell Res. Ther. 2018, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Romao, M.; Weel, I.C.; Lifshitz, S.J.; Peracoli, M.T. Elevated hyaluronan and extracellular matrix metalloproteinase inducer levels in women with preeclampsia. Arch. Gynecol. Obstet. 2014, 289, 575–579. [Google Scholar] [CrossRef]
- Chen, C.P.; Chen, L.F.; Yang, S.R.; Chen, C.Y.; Ko, C.C.; Chang, G.D.; Chen, H. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol. Reprod. 2008, 79, 815–823. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wang, R.; Yang, X.; Lu, X.Y.; Zhang, Q.; Wang, Y.L.; Wang, H.; Zhu, C.; Lin, H.Y.; Wang, H. The cAMP-responsive element binding protein (CREB) transcription factor regulates furin expression during human trophoblast syncytialization. Placenta 2014, 35, 907–918. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, T.; Zhang, B.H.; Lv, X.Y.; Lin, H.Y.; Zhu, C.; Xue, L.Q.; Wang, H. The proprotein convertase furin in human trophoblast: Possible role in promoting trophoblast cell migration and invasion. Placenta 2009, 30, 929–938. [Google Scholar] [CrossRef]
- Schwartz, D.A. An Analysis of 38 Pregnant Women with COVID-19, Their Newborn Infants, and Maternal-Fetal Transmission of SARS-CoV-2: Maternal Coronavirus Infections and Pregnancy Outcomes. Arch. Pathol. Lab. Med. 2020, 144, 799–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Algarroba, G.N.; Rekawek, P.; Vahanian, S.A.; Khullar, P.; Palaia, T.; Peltier, M.R.; Chavez, M.R.; Vintzileos, A.M. Visualization of SARS-CoV-2 virus invading the human placenta using electron microscopy. Am. J. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hosier, H.; Farhadian, S.F.; Morotti, R.A.; Deshmukh, U.; Lu-Culligan, A.; Campbell, K.H.; Yasumoto, Y.; Vogels, C.B.; Casanovas-Massana, A.; Vijayakumar, P.; et al. SARS-CoV-2 infection of the placenta. J. Clin. Investig. 2020. [Google Scholar] [CrossRef]
- Schoenmakers, S.; Snijder, P.; Verdijk, R.; Kuiken, T.; Kamphuis, S.; Koopman, L.; Krasemann, T.; Rousian, M.; Broekhuizen, M.; Steegers, E.; et al. SARS-CoV-2 placental infection and inflammation leading to fetal distress and neonatal multi-organ failure in an asymptomatic woman. medRxiv 2020. [Google Scholar] [CrossRef]
- Patane, L.; Morotti, D.; Giunta, M.R.; Sigismondi, C.; Piccoli, M.G.; Frigerio, L.; Mangili, G.; Arosio, M.; Cornolti, G. Vertical transmission of COVID-19: SARS-CoV-2 RNA on the fetal side of the placenta in pregnancies with COVID-19 positive mothers and neonates at birth. Am. J. Obstet. Gynecol. MFM 2020. [Google Scholar] [CrossRef]
- Buonsenso, D.; Costa, S.; Sanguinetti, M.; Cattani, P.; Posteraro, B.; Marchetti, S.; Carducci, B.; Lanzone, A.; Tamburrini, E.; Vento, G.; et al. Neonatal Late Onset Infection with Severe Acute Respiratory Syndrome Coronavirus 2. Am. J. Perinatol. 2020. [Google Scholar] [CrossRef]
- Penfield, C.A.; Brubaker, S.G.; Limaye, M.A.; Lighter, J.; Ratner, A.J.; Thomas, K.M.; Meyer, J.; Roman, A.S. Detection of SARS-COV-2 in Placental and Fetal Membrane Samples. Am. J. Obstet. Gynecol. MFM 2020. [Google Scholar] [CrossRef]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef]
- Zamaniyan, M.; Ebadi, A.; Aghajanpoor Mir, S.; Rahmani, Z.; Haghshenas, M.; Azizi, S. Preterm delivery in pregnant woman with critical COVID-19 pneumonia and vertical transmission. Prenat. Diagn. 2020. [Google Scholar] [CrossRef]
- Baud, D.; Greub, G.; Favre, G.; Gengler, C.; Jaton, K.; Dubruc, E.; Pomar, L. Second-Trimester Miscarriage in a Pregnant Woman With SARS-CoV-2 Infection. JAMA 2020. [Google Scholar] [CrossRef]
- Ferraiolo, A.; Barra, F.; Kratochwila, C.; Paudice, M.; Vellone, V.G.; Godano, E.; Varesano, S.; Noberasco, G.; Ferrero, S.; Arioni, C. Report of Positive Placental Swabs for SARS-CoV-2 in an Asymptomatic Pregnant Woman with COVID-19. Medicina 2020, 56, 306. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Guo, J.; Wang, C.; Luo, F.; Yu, X.; Zhang, W.; Li, J.; Zhao, D.; Xu, D.; Gong, Q.; et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020, 395, 809–815. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Guo, J.; Fan, C.; Juan, J.; Yu, X.; Li, J.; Feng, L.; Li, C.; Chen, H.; Qiao, Y.; et al. Coronavirus disease 2019 in pregnant women: A report based on 116 cases. Am. J. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, J.; Li, W.; Zhou, Z.; Liu, S.; Rong, Z. Clinical characteristics of 19 neonates born to mothers with COVID-19. Front. Med. 2020, 14, 193–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Tian, J.; He, S.; Zhu, C.; Wang, J.; Liu, C.; Yang, J. Possible Vertical Transmission of SARS-CoV-2 From an Infected Mother to Her Newborn. JAMA 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Xu, C.; Fan, J.; Tang, Y.; Deng, Q.; Zhang, W.; Long, X. Antibodies in Infants Born to Mothers With COVID-19 Pneumonia. JAMA 2020. [Google Scholar] [CrossRef]
- Walker, K.F.; O’Donoghue, K.; Grace, N.; Dorling, J.; Comeau, J.L.; Li, W.; Thornton, J.G. Maternal transmission of SARS-COV-2 to the neonate, and possible routes for such transmission: A systematic review and critical analysis. BJOG 2020. [Google Scholar] [CrossRef]
- Poon, L.C.; Yang, H.; Kapur, A.; Melamed, N.; Dao, B.; Divakar, H.; McIntyre, H.D.; Kihara, A.B.; Ayres-de-Campos, D.; Ferrazzi, E.M.; et al. Global interim guidance on coronavirus disease 2019 (COVID-19) during pregnancy and puerperium from FIGO and allied partners: Information for healthcare professionals. Int. J. Gynaecol. Obstet. 2020, 149, 273–286. [Google Scholar] [CrossRef]
- Egloff, C.; Vauloup-Fellous, C.; Picone, O.; Mandelbrot, L.; Roques, P. Evidence and possible mechanisms of rare maternal-fetal transmission of SARS-CoV-2. J. Clin. Virol. 2020, 128, 104447. [Google Scholar] [CrossRef]
- Trippella, G.; Ciarcia, M.; Ferrari, M.; Buzzatti, C.; Maccora, I.; Azzari, C.; Dani, C.; Galli, L.; Chiappini, E. COVID-19 in Pregnant Women and Neonates: A Systematic Review of the Literature with Quality Assessment of the Studies. Pathogens 2020, 9, 485. [Google Scholar] [CrossRef]
- Chawanpaiboon, S.; Vogel, J.P.; Moller, A.B.; Lumbiganon, P.; Petzold, M.; Hogan, D.; Landoulsi, S.; Jampathong, N.; Kongwattanakul, K.; Laopaiboon, M.; et al. Global, regional, and national estimates of levels of preterm birth in 2014: A systematic review and modelling analysis. Lancet Glob. Health 2019, 7, e37–e46. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Gao, J.; Luo, X.; Feng, L.; Liu, W.; Chen, J.; Benachi, A.; De Luca, D.; Chen, L. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vertical Transmission in Neonates Born to Mothers With Coronavirus Disease 2019 (COVID-19) Pneumonia. Obstet. Gynecol. 2020. [Google Scholar] [CrossRef]
- Khan, S.; Jun, L.; Nawsherwan; Siddique, R.; Li, Y.; Han, G.; Xue, M.; Nabi, G.; Liu, J. Association of COVID-19 with pregnancy outcomes in health-care workers and general women. Clin. Microbiol. Infect. 2020, 26, 788–790. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Xu, G.; Yang, Y.; Tao, Y.; Pian-Smith, M.; Madhavan, V.; Xie, Z.; Zhang, J. Evidence of mother-to-newborn infection with COVID-19. Br. J. Anaesth. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, L.; Chen, L.; Liu, W.; Cao, Y.; Zhang, J.; Feng, L. A case report of neonatal COVID-19 infection in China. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Yu, N.; Li, W.; Kang, Q.; Xiong, Z.; Wang, S.; Lin, X.; Liu, Y.; Xiao, J.; Liu, H.; Deng, D.; et al. Clinical features and obstetric and neonatal outcomes of pregnant patients with COVID-19 in Wuhan, China: A retrospective, single-centre, descriptive study. Lancet Infect. Dis. 2020, 20, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Xia, S.; Yuan, W.; Yan, K.; Xiao, F.; Shao, J.; Zhou, W. Neonatal Early-Onset Infection With SARS-CoV-2 in 33 Neonates Born to Mothers With COVID-19 in Wuhan, China. JAMA Pediatr. 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.J.; Yu, X.J.; Fu, T.; Liu, Y.; Jiang, Y.; Yang, B.X.; Bi, Y. Novel Coronavirus Infection in Newborn Babies Under 28 Days in China. Eur. Respir. J. 2020. [Google Scholar] [CrossRef] [Green Version]
- Alzamora, M.C.; Paredes, T.; Caceres, D.; Webb, C.M.; Valdez, L.M.; La Rosa, M. Severe COVID-19 during Pregnancy and Possible Vertical Transmission. Am. J. Perinatol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ferrazzi, E.; Frigerio, L.; Savasi, V.; Vergani, P.; Prefumo, F.; Barresi, S.; Bianchi, S.; Ciriello, E.; Facchinetti, F.; Gervasi, M.T.; et al. Vaginal delivery in SARS-CoV-2-infected pregnant women in Northern Italy: A retrospective analysis. BJOG 2020. [Google Scholar] [CrossRef]
- Kirtsman, M.; Diambomba, Y.; Poutanen, S.M.; Malinowski, A.K.; Vlachodimitropoulou, E.; Parks, W.T.; Erdman, L.; Morris, S.K.; Shah, P.S. Probable congenital SARS-CoV-2 infection in a neonate born to a woman with active SARS-CoV-2 infection. CMAJ 2020. [Google Scholar] [CrossRef]
- Mehta, H.; Ivanovic, S.; Cronin, A.; VanBrunt, L.; Mistry, N.; Miller, R.; Yodice, P.; Rezai, F. Novel coronavirus-related acute respiratory distress syndrome in a patient with twin pregnancy: A case report. Case Rep. Womens Health 2020, e00220. [Google Scholar] [CrossRef]
- Piersigilli, F.; Carkeek, K.; Hocq, C.; van Grambezen, B.; Hubinont, C.; Chatzis, O.; Van der Linden, D.; Danhaive, O. COVID-19 in a 26-week preterm neonate. Lancet Child. Adolesc. Health 2020, 4, 476–478. [Google Scholar] [CrossRef]
- Blumberg, D.A.; Underwood, M.A.; Hedriana, H.L.; Lakshminrusimha, S. Vertical Transmission of SARS-CoV-2: What is the Optimal Definition? Am. J. Perinatol. 2020, 37, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, B.; Luo, D.J.; Li, X.; Yang, F.; Zhao, Y.; Nie, X.; Huang, B.X. Pregnancy with new coronavirus infection: Clinical characteristics and placental pathological analysis of three cases. Zhonghua Bing Li Xue Za Zhi 2020, 49, 418–423. [Google Scholar] [CrossRef]
- Baergen, R.N.; Heller, D.S. Placental Pathology in Covid-19 Positive Mothers: Preliminary Findings. Pediatr. Dev. Pathol. 2020, 23, 177–180. [Google Scholar] [CrossRef]
- Mulvey, J.J.; Magro, C.M.; Ma, L.X.; Nuovo, G.J.; Baergen, R.N. Analysis of complement deposition and viral RNA in placentas of COVID-19 patients. Ann. Diagn. Pathol. 2020, 46, 151530. [Google Scholar] [CrossRef] [PubMed]
- Ernst, L.M. Maternal vascular malperfusion of the placental bed. APMIS 2018, 126, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, W.T.; Catov, J.M. The Placenta as a Window to Maternal Vascular Health. Obstet. Gynecol. Clin. N. Am. 2020, 47, 17–28. [Google Scholar] [CrossRef]
- Roberts, D.J.; Post, M.D. The placenta in pre-eclampsia and intrauterine growth restriction. J. Clin. Pathol. 2008, 61, 1254–1260. [Google Scholar] [CrossRef]
- Shanes, E.D.; Mithal, L.B.; Otero, S.; Azad, H.A.; Miller, E.S.; Goldstein, J.A. Placental Pathology in COVID-19. Am. J. Clin. Pathol. 2020, 154, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Heida, K.Y.; Velthuis, B.K.; Oudijk, M.A.; Reitsma, J.B.; Bots, M.L.; Franx, A.; van Dunne, F.M.; Dutch Guideline Development Group on Cardiovascular Risk Management after Reproductive Disorders. Cardiovascular disease risk in women with a history of spontaneous preterm delivery: A systematic review and meta-analysis. Eur. J. Prev. Cardiol. 2016, 23, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Steegers, E.A.; von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Pre-eclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Contro, E.; deSouza, R.; Bhide, A. Chronic intervillositis of the placenta: A systematic review. Placenta 2010, 31, 1106–1110. [Google Scholar] [CrossRef]
- Ng, W.F.; Wong, S.F.; Lam, A.; Mak, Y.F.; Yao, H.; Lee, K.C.; Chow, K.M.; Yu, W.C.; Ho, L.C. The placentas of patients with severe acute respiratory syndrome: A pathophysiological evaluation. Pathology 2006, 38, 210–218. [Google Scholar] [CrossRef]
- Wang, C.; Xie, J.; Zhao, L.; Fei, X.; Zhang, H.; Tan, Y.; Nie, X.; Zhou, L.; Liu, Z.; Ren, Y.; et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 2020, 57, 102833. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Donker, R.B.; Mouillet, J.F.; Chu, T.; Bayer, A.; Ouyang, Y.; Wang, T.; Stolz, D.B.; Sarkar, S.N.; Morelli, A.E.; et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12048–12053. [Google Scholar] [CrossRef] [Green Version]
- Ander, S.E.; Diamond, M.S.; Coyne, C.B. Immune responses at the maternal-fetal interface. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Liu, S.; Diao, L.; Huang, C.; Li, Y.; Zeng, Y.; Kwak-Kim, J.Y.H. The role of decidual immune cells on human pregnancy. J. Reprod. Immunol. 2017, 124, 44–53. [Google Scholar] [CrossRef]
- Carlino, C.; Stabile, H.; Morrone, S.; Bulla, R.; Soriani, A.; Agostinis, C.; Bossi, F.; Mocci, C.; Sarazani, F.; Tedesco, F.; et al. Recruitment of circulating NK cells through decidual tissues: A possible mechanism controlling NK cell accumulation in the uterus during early pregnancy. Blood 2008, 111, 3108–3115. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhu, X.Y.; Du, M.R.; Li, D.J. Human trophoblasts recruited T lymphocytes and monocytes into decidua by secretion of chemokine CXCL16 and interaction with CXCR6 in the first-trimester pregnancy. J. Immunol. 2008, 180, 2367–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erlebacher, A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 2013, 31, 387–411. [Google Scholar] [CrossRef]
- Magatti, M.; Stefani, F.R.; Papait, A.; Cargnoni, A.; Masserdotti, A.; Silini, A.R.; Parolini, O. Perinatal Mesenchymal Stromal Cells and Their Possible Contribution to Fetal-Maternal Tolerance. Cells 2019, 8, 1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louwen, F.; Ritter, A.; Kreis, N.N.; Yuan, J. Insight into the development of obesity: Functional alterations of adipose-derived mesenchymal stem cells. Obes. Rev. 2018, 19, 888–904. [Google Scholar] [CrossRef] [PubMed]
- Reyes, L.; Golos, T.G. Hofbauer Cells: Their Role in Healthy and Complicated Pregnancy. Front. Immunol. 2018, 9, 2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loegl, J.; Hiden, U.; Nussbaumer, E.; Schliefsteiner, C.; Cvitic, S.; Lang, I.; Wadsack, C.; Huppertz, B.; Desoye, G. Hofbauer cells of M2a, M2b and M2c polarization may regulate feto-placental angiogenesis. Reproduction 2016, 152, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karaman, O.M.; Ivanchenko, A.V.; Chekhun, V.F. Macrophages—a perspective target for antineoplastic immunotherapy. Exp. Oncol. 2019, 41, 282–290. [Google Scholar] [CrossRef]
- Roopenian, D.C.; Akilesh, S. FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 2007, 7, 715–725. [Google Scholar] [CrossRef]
- Maltepe, E.; Fisher, S.J. Placenta: The forgotten organ. Annu. Rev. Cell. Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef]
- Koga, K.; Izumi, G.; Mor, G.; Fujii, T.; Osuga, Y. Toll-like receptors at the maternal-fetal interface in normal pregnancy and pregnancy complications. Am. J. Reprod. Immunol. 2014, 72, 192–205. [Google Scholar] [CrossRef]
- Levy, O. Innate immunity of the newborn: Basic mechanisms and clinical correlates. Nat. Rev. Immunol. 2007, 7, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, V.M. The role of the Nod-like receptor family in trophoblast innate immune responses. J. Reprod. Immunol. 2011, 88, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.H.; Wang, Y.N.; Chang, Q.Y.; Ma, P.; Hu, Y.; Cao, X. Type III Interferons in Viral Infection and Antiviral Immunity. Cell Physiol. Biochem. 2018, 51, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [Green Version]
- Wells, A.I.; Coyne, C.B. Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol. 2018, 39, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Kotenko, S.V.; Gallagher, G.; Baurin, V.V.; Lewis-Antes, A.; Shen, M.; Shah, N.K.; Langer, J.A.; Sheikh, F.; Dickensheets, H.; Donnelly, R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003, 4, 69–77. [Google Scholar] [CrossRef]
- Hamming, O.J.; Terczynska-Dyla, E.; Vieyres, G.; Dijkman, R.; Jorgensen, S.E.; Akhtar, H.; Siupka, P.; Pietschmann, T.; Thiel, V.; Hartmann, R. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 2013, 32, 3055–3065. [Google Scholar] [CrossRef]
- de Weerd, N.A.; Nguyen, T. The interferons and their receptors--distribution and regulation. Immunol. Cell Biol. 2012, 90, 483–491. [Google Scholar] [CrossRef]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Bramley, J.C.; Morosky, S.; Marques, E.T., Jr.; Cherry, S.; Sadovsky, Y.; Coyne, C.B. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe 2016, 19, 705–712. [Google Scholar] [CrossRef] [Green Version]
- Jagger, B.W.; Miner, J.J.; Cao, B.; Arora, N.; Smith, A.M.; Kovacs, A.; Mysorekar, I.U.; Coyne, C.B.; Diamond, M.S. Gestational Stage and IFN-lambda Signaling Regulate ZIKV Infection In Utero. Cell Host Microbe 2017, 22, 366–376.e3. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Mouillet, J.F.; Coyne, C.B.; Sadovsky, Y. Review: Placenta-specific microRNAs in exosomes—Good things come in nano-packages. Placenta 2014, 35, S69–S73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Bayer, A.; Delorme-Axford, E.; Sleigher, C.; Frey, T.K.; Trobaugh, D.W.; Klimstra, W.B.; Emert-Sedlak, L.A.; Smithgall, T.E.; Kinchington, P.R.; Vadia, S.; et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am. J. Obstet. Gynecol. 2015, 212, 71.e1–71.e8. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Bayer, A.; Sadovsky, Y.; Coyne, C.B. Autophagy as a mechanism of antiviral defense at the maternal-fetal interface. Autophagy 2013, 9, 2173–2174. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [Green Version]
- Shoji-Kawata, S.; Levine, B. Autophagy, antiviral immunity, and viral countermeasures. Biochim. Biophys. Acta 2009, 1793, 1478–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.; Sabit, H. microRNA-Based Vaccination and Treatment for COVID-19. Curr. Trends Vaccines Vaccinol. 2020, 3. [Google Scholar] [CrossRef]
- Schmitz, M.L.; Kracht, M.; Saul, V.V. The intricate interplay between RNA viruses and NF-kappaB. Biochim. Biophys. Acta 2014, 1843, 2754–2764. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Lingappan, K. NF-kappaB in Oxidative Stress. Curr. Opin. Toxicol. 2018, 7, 81–86. [Google Scholar] [CrossRef]
- Armistead, B.; Kadam, L.; Drewlo, S.; Kohan-Ghadr, H.R. The Role of NFkappaB in Healthy and Preeclamptic Placenta: Trophoblasts in the Spotlight. Int. J. Mol. Sci. 2020, 21, 1775. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.S.; Ghosh, S. Regulation of NF-kappaB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef] [Green Version]
- Schutze, S.; Wiegmann, K.; Machleidt, T.; Kronke, M. TNF-induced activation of NF-kappa B. Immunobiology 1995, 193, 193–203. [Google Scholar] [CrossRef]
- Batra, S.; Balamayooran, G.; Sahoo, M.K. Nuclear factor-kappaB: A key regulator in health and disease of lungs. Arch. Immunol. Ther. Exp. 2011, 59, 335–351. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Nieto-Torres, J.L.; Regla-Nava, J.A.; Jimenez-Guardeno, J.M.; Fernandez-Delgado, R.; Fett, C.; Castano-Rodriguez, C.; Perlman, S.; Enjuanes, L. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014, 88, 913–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dosch, S.F.; Mahajan, S.D.; Collins, A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009, 142, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Cardenas, I.; Abrahams, V.; Guller, S. Inflammation and pregnancy: The role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 2011, 1221, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Haider, S.; Meinhardt, G.; Saleh, L.; Kunihs, V.; Gamperl, M.; Kaindl, U.; Ellinger, A.; Burkard, T.R.; Fiala, C.; Pollheimer, J.; et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Rep. 2018, 11, 537–551. [Google Scholar] [CrossRef] [Green Version]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, M.S.; Strawn, W.B.; Groban, L.; Yamaleyeva, L.M.; Chappell, M.C.; Horta, C.; Atkins, K.; Firmes, L.; Gurley, S.B.; Brosnihan, K.B. Angiotensin-converting enzyme 2 deficiency is associated with impaired gestational weight gain and fetal growth restriction. Hypertension 2011, 58, 852–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Reference | Method/Datasets | Placental Tissue/Patient Information | Receptors/Proteases | Main Results |
|---|---|---|---|---|
| Transmission Possible | ||||
| Valdés et al. [58] | IHC | 11 early pregnancy failures (5 miscarriages, 9.5 ± 2.2 w; 6 ectopic pregnancies, 7.4 ± 1.9 w), 15 normotensive (38.7 ± 0.9 w) and 10 preeclamptic placentas (35 ± 2.9 w) | ACE2 | ACE2 is expressed in placental villi (STB, vCTBs, vascular smooth muscle cells, endothelium) and in the maternal stroma (EVTs, decidual cells). ACE2 expression is comparable between normal term and PE, except for increased ACE2 in umbilical arterial endothelium in PE. |
| Pringle et al. [59] | mRNA expression IHC | Early gestation placenta (6–16 w, n = 14 for IHC, n = 22–26 for mRNA, elective terminations) and term (37-41 w, n = 7–9 for mRNA, CS) | ACE2 | ACE2 mRNA is highest in early gestational samples with a sharp decline at term. ACE2 protein is abundant in the STB and villous stroma, and in a lesser extent in vCTBs in early gestation placenta; not mentioned for term. |
| Li et al. [60] | scRNA-seq E-MTAB-6701 [61], GSE89497 [62] and datasets of fetal heart [63], liver or kidney [64] | 11 deciduas and 5 placentas from 6–14 w [61]; 1st trimester: villi from 8 w placentas and 2nd trimester: 24 w placenta for EVTs [62] | ACE2 TMPRSS2 | ACE2 is highly abundant in decidual stromal cells and perivascular cells, and in vCTBs and the STB. TMPRSS2 is expressed in vCTBs and epithelial glandular cells, and low in the STB. ACE2 expression is significantly increased in EVTs at later stage of pregnancy (24 w). TMPRSS2 has a similar dynamic alteration in the STB. ACE2 and TMPRSS2 are co-expressed in vCTBs, the STB and EVTs. ACE2 is also expressed in specific cell types of human fetal heart and liver. |
| Ashray et al. [65] | scRNA-seq GSE89497 [62] and [66] | 1st trimester: villi from 8 w placentas and 2nd trimester: 24 w placenta for EVTs [62]; 1st trimester (6–11 w) villi (n = 8) and decidua (n = 6) [66] | ACE2 TMPRSS2 CD147 CTSL FURIN | ACE2 and TMPRSS2 are co-expressed in 14% STB in 1st trimester placenta and in 15% EVTs in 2nd trimester placenta. 18% vCTBs express ACE2. CD147 and CTSL are abundant in almost all EVTs, the STB, vCTBs and villous stromal cells. The ACE2+/TMPRSS2+ STB and EVTs abundantly express FURIN as well as mRNA for proteins involved in viral budding and replication. |
| Colaco et al. [67] | scRNA-seq of human embryos [68,69] | Human pre-implantation embryos and hESCs [68]; transcriptome signature for human pre-implantation epiblast [69] | ACE2 TMPRSS2 CD147 CTSL DPP4 | ACE2 and TMPRSS2 are co-expressed in a proportion of epiblast cells. CD147, CTSL and DPP4 are expressed in all stages of embryonic development. The cells of the epiblast express genes involved in viral endocytosis and replication. Cells of the blastocyst also express DPP4. In ACE2+/TMPRSS2+ cells of the epiblast, biological processes of placental development and viral entry in host cells are enriched. |
| Transmission Not Likely | ||||
| Zheng et al. [70] | scRNA-seq E-MTAB-6678 [61] | 11 deciduas and 5 placentas from 6–14 w | ACE2 | The majority of ACE2-expressing cells are perivascular cells. Its expression is very low in the STB, decidual stromal cells and epithelial glandular cells. |
| Sungnak et al. [71] | scRNA-seq E-MTAB-6701 [61] | 11 deciduas and 5 placentas from 6–14 w | ACE2 TMPRSS2 | ACE2 expression is noticeable in certain cell types in the placenta or the decidua (the STB, vCTBs or decidual perivascular cells, decidual stromal cells) w/o TMPRSS2. |
| Pique-Regi et al. [72] | scRNA-seq E-MTAB-6701 [61], NIH dbGAP phs001886.v1.p1 [73], newly generated scRNA-seq/snRNA-seq data [72], microarray datasets E-TABM-577 [74,75] | 11 deciduas and 5 placentas from 6-14 w [61]; 3rd trimester placenta: term (38-40 w, n = 6) or preterm (33–35 w, n = 3), 66.67% CS [73]. Placenta accreta at 18 w and 32 patients at 3rd trimester placenta (32.9–39.1 w, 59.4% CS) [72]. 3rd trimester placenta: spontaneous labor at term w/o (n=10) and with villitis of unknown etiology (n=10) [74]; PE/SGA/PE+SGA samples (n = 28, ~34.2 w) [75]. | ACE2 TMPRSS2 CD147 CTSL FURIN | Very few cells co-express ACE2 and TMPRSS2 in any of the three trimesters, also minimally detected in chorioamniotic membranes. Placenta and chorioamniotic membranes express high levels of CD147 throughout pregnancy. CTSL and FURIN are highly abundant in placental tissue throughout gestation. The analyses of bulk gene expression data revealed that ACE2 is detected in most samples, while TMPRSS2 is largely undetected. |
| Constantino et al. [76] | Affymetrix microarray dataset GSE9984 [77] and scRNA-seq E-MTAB-6701 [61] | Villous trophoblast tissues of 1st trimester (45–59 days, n = 4), and 2nd trimester (109–115 days, n = 4) from uncomplicated elective terminations and 3rd trimester (n = 4) from term, CS [77]; early maternal-fetal interface (11 deciduas and 5 placentas from 6–14 w) [61]. | ACE2 TMPRSS2 DPP4 CTSL | Low levels of ACE2 and TMPRSS2, but villous trophoblast cells co-express high levels of DPP4 and CTSL throughout gestation. DPP4 and CTSL are highly co-expressed in the STB, vCTB and EVTs. |
| Reference | Patient Information | Material/Method | Positive Neonates | Possible Vertical Transmission |
|---|---|---|---|---|
| Algarroba et al. (NY, USA) [94] | 28 w + 4 d, 40 y old, CS, severe infection | Placental evaluation using TEM | 0 | Viral particle detected in the STB and villous fibroblasts. |
| Alzamora et al. (Lima, Peru) [121] | 33 w, 41 y old, diabetes, BMI 35 kg/m2, CS, severe infection | RT-PCR | 1 | Neonatal isolation, positive NP swab 16 h and 48 h postpartum (pp). |
| Buonsenso et al. (Rome, Italy) [98] | Case 1: 38 w + 3 d, 42 y old, CS, symptomatic COVID-19 with cough Case 2: 35 w+5 d, 38 y old, CS, symptomatic with cough, dyspnea and fever | Serology RT-PCR | 1 | Case 1: 24 h pp: slightly elevated IgG; 24 h and 3 d pp: negative NP swab; 15 d pp: positive NP swab, in-hospital separation; at home (5 d) breastfeeding with mask; late-onset infection. Case 2: NP swabs negative, RT-PCR positive placenta and umbilical blood; positive breast milk (3 out of 5 samples). |
| Dong et al. (China) [107] | 34 w + 2 d, 29 y, CS, symptomatic | Serology/CLIA | (1) * | Elevated IgM and IgG, IL-6 and IL-10; 2 h pp; elevated IgM antibody level suggests that the neonate was infected in utero. Negative RT-PCR. |
| Ferraiolo et al. (Genoa, Italy) [103] | 38 w + 3 d, 30 y old, CS, asymptomatic woman | RT-PCR | 0 | Placental swabs obtained from the fetal surface proximate to the umbilical cord were positive for SARS-CoV-2 RNA. |
| Ferrazzi et al. (Italy) [122] | 42 cases, 30 term, mean age 32.9 y old (range 21–44 y), 18 CS/24 VD, 6 with gestational diabetes, generally mild to moderate symptoms | RT-PCR | 3 | 2 newborns had positive NP swabs on day 1 and 3, breastfeeding, skin-to-skin contact allowed. 1 newborn (VD) had a positive test with respiratory symptoms on day 3, without breastfeeding. |
| Hosier et al. (Connecticut, USA) [95] | 22 w, 35 y old, severe PE, elevated transaminases, and low platelets, placental abruption with termination of pregnancy, symptomatic | RT-PCR, sequencing of SARS-CoV-2 RNA (placenta) IHC, ISH and TEM (placenta) | 0 | SARS-CoV-2-RNA positive placenta and umbilical cord; fetal lungs, heart and kidney were negative. IHC for SARS-CoV-2 spike protein: predominantly in the STB, confirmed with ISH. TEM: virus particles within the cytosol of placental cells (vCTB, STB, fibroblasts). |
| Hu. et al. (China) [114] | 7 cases, 38 w + 2 d to 41 w + 2 d, 30–34 y old, 6 CS/1 VD | RT-PCR | 1 | 1 NP swab positive 36 h postnatal (CS), neonatal isolation for 14 d, formula-fed, symptomatic mother. |
| Khan et al. (China) [115] | 17 cases, mean 38.1 w (35 w + 5 d-41 w), mean 29.29 y old (24—34 y), CS | RT-PCR | 2 | 2 neonates with positive NP swab 24 h postnatal, with 1 developing pneumonia; 4 neonates with pneumonia and negative NP swab. |
| Kirtsman et al. (Toronto, Canada)[123] | 35 w + 5 d, 40 y old, CS, maternal familial neutropenia and gestational diabetes, symptomatic | RT-PCR | 1 | NP swab positive on day of birth (no skin-to-skin contact), day 2 and day 7. Plasma positive on day 4 and stool positive on day 7; breastfeeding otherwise keeping a distance; breast milk positive. |
| Knight et al. (UK) [9] | 244 cases, late second and third trimester; 4VD, 8CS | RT-PCR | 12 | 12 infants (5%) positive for SARS-CoV-2 RNA, 6 of these infants within the first 12 h after birth. |
| Mehta et al. (New Jersey, USA) [124] | dichorionic twins, 28 w, 39 y old, IVF, CS, symptomatic | RT-PCR | 1 | One twin tested positive 72 h after birth. |
| Patané et al. (Bergamo, Italy; * possible duplication with Ferrazzi et al.) [97] | 22 cases, case 1: 37 w + 6 d, VD, symptomatic case 2: 35 w + 1 d, CS, symptomatic | RT-PCR ISH | (2) * | 2 neonates with positive NP swab. Case 1: positive at 24 h and on day 7, NP swab with skin-to-skin contact and breastfeeding. Case 2: positive NP swab on day 7, separation. ISH: positive dots for SARS-CoV-2 spike protein mRNA in the STB of both placentas. |
| Penfield et al. (NY, USA) [99] | 11 cases, 26 w + 5 d–41 w + 3 d, 22-40 y old, symptomatic | RT-PCR | 0 | 1 positive placental swab (CS, severe COVID-19) and 2 positive membrane swabs (between the amnion and chorion membrane; CS, critical COVID-19). |
| Piersigilli et al. (Brussels, Belgium) [125] | 26 w + 4 d, PE and HELLP, CS, symptomatic the day after delivery | RT-PCR | 1 | Positive NP swab on day 7. Possible pp infection. |
| Schoenmakers et al. [96], (Rotterdam, The Netherlands) | 3rd trimester, obesity and gestational diabetes, CS, asymptomatic at presentation | RT-PCR TEM IHC ISH | 0 | Positive placenta at maternal and fetal side (RT-PCR). SARS-CoV-2 particles were detected in the STB (TEM, IHC, ISH). Fetal distress but negative for SARS-CoV-2. |
| Sun et al. (China) [116] | 3 cases: 30 w + 5 d–37w; 28-30 y old, CS, symptomatic COVID-19 | RT-PCR Chest CT scan | 1 | 1 positive NP swab on day 6. 1 CT on day 6 showed findings suggestive of COVID-19. |
| Vivanti et al. (Paris, France) [100] | 35 w + 5 d, 23 y old, CS, symptomatic | RT-PCR | 1 | Amniotic fluid, placenta, neonatal blood and non-bronchoscopic bronchoalveolar lavage fluid were positive for E and S genes of SARS-CoV-2. NP and rectal swabs positive: 1 h, 3 d and 18 d postnatal; formula feeding; neonatal viremia. |
| Wang et al. (China) [117] | 40 w, 34 y old, CS, low-grade fever | RT-PCR | 1 | Positive NP swab 36 h pp; cord blood, placenta and breast milk were negative. |
| Yu et al. (China) [118] * | 7 cases, 39 w + 1 d (37–41 w + 2 d), 32 y old (29–34 y), CS, symptomatic | RT-PCR | 1 | 1 neonate was infected with SARS-CoV-2 36 h after birth (throat swab); placenta and cord blood negative. |
| Zamaniyan et al. (Sari, Iran) [101] | 32 w, 22 y old, CS, severe COVID-19 and controlled hypothyroidism | RT-PCR | 1 | Positive amniotic fluid and repeated neonatal NP swabs on day 1 and 7 pp; isolated without breastfeeding; maternal death. |
| Zeng et al. (China) [108] | 6 cases, 3rd trimester, CS, mild COVID-19 | Serology/ CLIA | (2) * | IgG elevated in 5 neonates, IgM in 2. Infants were isolated from mothers. Negative RT-PCR. |
| Zeng et al. (China) [119] | 33 cases, 3 positives: 31 w + 2 d, 40 w, 40 w + 2 d; CS, symptomatic | RT-PCR | 3 | NP/anal swabs were positive for SARS-CoV-2 on day 2, 4 and negative on day 6 or 7, respectively. |
| Zhang et al. (China) [120] | 4 cases, CS, symptomatic (3 mothers with symptoms before and 1 after delivery) | RT-PCR | 4 | Infections detected between 30 h, 5 and 17 days of life (NP and anal swabs). Mild symptoms. No mother-child contact/breastfeeding in 3 cases. |
| Total | 35 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreis, N.-N.; Ritter, A.; Louwen, F.; Yuan, J. A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2. Cells 2020, 9, 1777. https://doi.org/10.3390/cells9081777
Kreis N-N, Ritter A, Louwen F, Yuan J. A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2. Cells. 2020; 9(8):1777. https://doi.org/10.3390/cells9081777
Chicago/Turabian StyleKreis, Nina-Naomi, Andreas Ritter, Frank Louwen, and Juping Yuan. 2020. "A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2" Cells 9, no. 8: 1777. https://doi.org/10.3390/cells9081777
APA StyleKreis, N.-N., Ritter, A., Louwen, F., & Yuan, J. (2020). A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2. Cells, 9(8), 1777. https://doi.org/10.3390/cells9081777





