Atg8-Family Proteins—Structural Features and Molecular Interactions in Autophagy and Beyond
Abstract
:1. Introduction
2. Ubiquitin, UBLs, and Atg8-Family Proteins
2.1. Structural Overview
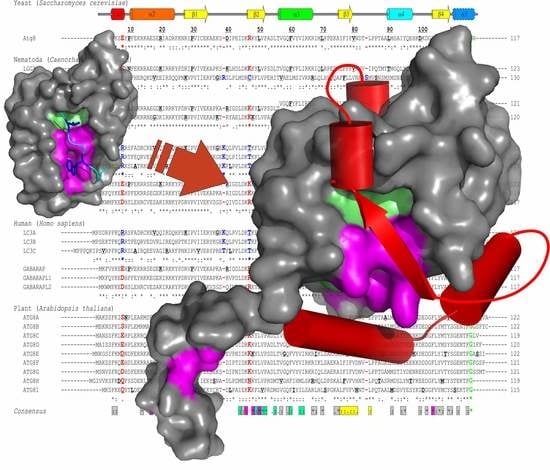
2.2. Lessons from Alignment of Atg8 Family Members
3. Interactions between Atg8/LC3/GABARAP Proteins and Their Binding Partners
3.1. The LIR Concept
3.2. Affinity and Selectivity of Interactions between Atg8/LC3/GABARAPs and LIR Motifs
3.3. Atg8/LC3/GABARAP Interactions: LIR and Beyond
3.3.1. Linear LIR-Like Motifs
3.3.2. Three-Dimensional (3D) Interacting Regions (3D LIRs)
3.3.3. Extended LIR Motifs: C-Terminal α-Helical Extensions
3.3.4. Extended LIR Motifs: N- and C-Terminal β-Stranded Extensions
3.3.5. α-Helical Interacting Regions
3.3.6. UIM-Like Interacting Regions
4. Emerging Atg8/LC3/GABARAP Interaction Motifs and Elements
4.1. The Anti-Parallel Intermolecular β-Sheet
4.2. N-Terminal α-Helical Subdomain Displacement
4.3. LIR-Based Atg8/LC3/GABARAP Superbinders or Combinatorial Binder
5. Therapeutic Exploitation of the LIR: Atg8/LC3/GABARAP Interactions
Funding
Acknowledgments
Conflicts of Interest
References
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell. Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Rogov, V.V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Tooze, S.A.; Yoshimori, T. The origin of the autophagosomal membrane. Nat. Cell Biol. 2010, 12, 831–835. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef] [Green Version]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Ohsumi, Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell Mol. Biol. 2016, 321, 29–88. [Google Scholar] [CrossRef] [Green Version]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR motif–crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, T.M.; Morano, K.A.; Scott, S.V.; Klionsky, D.J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995, 131, 591–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Alberti, A.; Michelet, X.; Djeddi, A.; Legouis, R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 2010, 6, 622–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juhász, G.; Érdi, B.; Sass, M.; Neufeld, T.P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007, 21, 3061–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.-i.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Karsli-Uzunbas, G.; Guo, J.Y.; Price, S.; Teng, X.; Laddha, S.V.; Khor, S.; Kalaany, N.Y.; Jacks, T.; Chan, C.S.; Rabinowitz, J.D.; et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014, 4, 914–927. [Google Scholar] [CrossRef] [Green Version]
- Cassidy, L.D.; Young, A.R.; Young, C.N.; Soilleux, E.J.; Fielder, E.; Weigand, B.M.; Lagnado, A.; Brais, R.; Ktistakis, N.T.; Wiggins, K.A.; et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef] [Green Version]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef]
- Torii, S.; Yamaguchi, H.; Nakanishi, A.; Arakawa, S.; Honda, S.; Moriwaki, K.; Nakano, H.; Shimizu, S. Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy. Nat. Commun. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tsuboyama, K.; Koyama-Honda, I.; Sakamaki, Y.; Koike, M.; Morishita, H.; Mizushima, N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016, 354, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, G.; Martens, S. Mechanisms of selective autophagy. J. Mol. Biol. 2016, 428, 1714–1724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, T.; Lamark, T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kumanomidou, T.; Sou, Y.-s.; Mizushima, T.; Ezaki, J.; Ueno, T.; Kominami, E.; Yamane, T.; Tanaka, K.; Komatsu, M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008, 283, 22847–22857. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.N.; Kumeta, H.; Nakatogawa, H.; Satoo, K.; Adachi, W.; Ishii, J.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 2008, 13, 1211–1218. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.-A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [Green Version]
- Jacomin, A.-C.; Samavedam, S.; Promponas, V.; Nezis, I.P. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016, 12, 1945–1953. [Google Scholar] [CrossRef] [Green Version]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, J.M.; Ye, Y.; Gebru, M.T.; Liu, Q.; Zhou, S.; Young, M.M.; Takahashi, Y.; Lin, Q.; Tian, F.; Wang, H.-G. Time-resolved FRET and NMR analyses reveal selective binding of peptides containing the LC3-interacting region to ATG8 family proteins. J. Biol. Chem. 2019, 294, 14033–14042. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.; Birgisdottir, Å.; Huber, J.; Kniss, A.; Dötsch, V.; Kirkin, V.; Rogov, V. Methods for studying interactions between Atg8/LC3/GABARAP and LIR-containing proteins. Methods Enzymol. 2017, 587, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.S.; Birgisdottir, Å.B.; Johansen, T. Use of Peptide Arrays for Identification and Characterization of LIR Motifs. Methods Mol. Biol. 2019, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.V.; Stolz, A.; Ravichandran, A.C.; Rios-Szwed, D.O.; Suzuki, H.; Kniss, A.; Löhr, F.; Wakatsuki, S.; Dötsch, V.; Dikic, I.; et al. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep. 2017, 18, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Schulman, B.A. Atomistic autophagy: The structures of cellular self-digestion. Cell 2014, 157, 300–311. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.N.; Ohsumi, Y.; Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010, 584, 1379–1385. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.; Dötsch, V.; Johansen, T.; Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Sora, V.; Kumar, M.; Maiani, E.; Lambrughi, M.; Tiberti, M.; Papaleo, E. Structure and dynamics in the ATG8 family from experimental to computational techniques. Front. Cell Dev. Biol. 2020, 8, 420. [Google Scholar] [CrossRef]
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. Structure of ubiquitin refined at 1.8 Åresolution. J. Mol. Biol. 1987, 194, 531–544. [Google Scholar] [CrossRef]
- Kirkin, V.; Dikic, I. Role of ubiquitin-and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 2007, 19, 199–205. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Suzuki, N.N.; Obara, K.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of Atg5· Atg16, a complex essential for autophagy. J. Biol. Chem. 2007, 282, 6763–6772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fracchiolla, D.; Sawa-Makarska, J.; Zens, B.; de Ruiter, A.; Zaffagnini, G.; Brezovich, A.; Romanov, J.; Runggatscher, K.; Kraft, C.; Zagrovic, B.; et al. Mechanism of cargo-directed Atg8 conjugation during selective autophagy. Elife 2016, 5, e18544. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.; Moharir, S.C.; Sailasree, S.P.; Sirohi, K.; Sudhakar, C.; Sarathi, D.P.; Lakshmi, B.J.; Buono, M.; Kumar, S.; Swarup, G. Optineurin promotes autophagosome formation by recruiting the autophagy-related Atg12-5-16L1 complex to phagophores containing the Wipi2 protein. J. Biol. Chem. 2018, 293, 132–147. [Google Scholar] [CrossRef] [Green Version]
- Lane, J.D.; Baines, K. The ATG5 Interactome Links Clathrin Vesicular Trafficking With The ATG8 Lipidation Machinery For Autophagosome Assembly. bioRxiv 2019, 769059. [Google Scholar] [CrossRef]
- Mohrlüder, J.; Hoffmann, Y.; Stangler, T.; Hänel, K.; Willbold, D. Identification of clathrin heavy chain as a direct interaction partner for the γ-aminobutyric acid type A receptor associated protein. Biochemistry 2007, 46, 14537–14543. [Google Scholar] [CrossRef]
- Radoshevich, L.; Murrow, L.; Chen, N.; Fernandez, E.; Roy, S.; Fung, C.; Debnath, J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010, 142, 590–600. [Google Scholar] [CrossRef] [Green Version]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [Green Version]
- Huber, J.; Obata, M.; Gruber, J.; Akutsu, M.; Löhr, F.; Rogova, N.; Güntert, P.; Dikic, I.; Kirkin, V.; Komatsu, M.; et al. An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5. Autophagy 2020, 16, 256–270. [Google Scholar] [CrossRef]
- Liu, X.-M.; Yamasaki, A.; Du, X.-M.; Coffman, V.C.; Ohsumi, Y.; Nakatogawa, H.; Wu, J.-Q.; Noda, N.N.; Du, L.-L. Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein. Elife 2018, 7, e41237. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Hayashi-Nishino, M.; Fukumoto, H.; Omori, H.; Yamamoto, A.; Noda, T.; Yoshimori, T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 2008, 19, 4651–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020, 48, D376–D382. [Google Scholar] [CrossRef] [PubMed]
- Coyle, J.E.; Qamar, S.; Rajashankar, K.R.; Nikolov, D.B. Structure of GABARAP in two conformations: Implications for GABAA receptor localization and tubulin binding. Neuron 2002, 33, 63–74. [Google Scholar] [CrossRef] [Green Version]
- Krichel, C.; Möckel, C.; Schillinger, O.; Huesgen, P.F.; Sticht, H.; Strodel, B.; Weiergräber, O.H.; Willbold, D.; Neudecker, P. Solution structure of the autophagy-related protein LC3C reveals a polyproline II motif on a mobile tether with phosphorylation site. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Stangler, T.; Mayr, L.M.; Willbold, D. Solution Structure of Human GABAA Receptor-associated Protein GABARAP: Implications for Biological Function and its Regulation. J. Biol. Chem. 2002, 277, 13363–13366. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Weidberg, H.; Shpilka, T.; Shvets, E.; Abada, A.; Shimron, F.; Elazar, Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell 2011, 20, 444–454. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.T.; Ji, J.; Dagda, R.K.; Jiang, J.F.; Tyurina, Y.Y.; Kapralov, A.A.; Tyurin, V.A.; Yanamala, N.; Shrivastava, I.H.; Mohammadyani, D.; et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013, 15, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Thielmann, Y.; Mohrlüder, J.; Koenig, B.W.; Stangler, T.; Hartmann, R.; Becker, K.; Höltje, H.D.; Willbold, D. An indole-binding site is a major determinant of the ligand specificity of the GABA type A receptor-associated protein GABARAP. ChemBioChem 2008, 9, 1767–1775. [Google Scholar] [CrossRef] [PubMed]
- Cook, W.J.; Jeffrey, L.C.; Kasperek, E.; Pickart, C.M. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J. Mol. Biol. 1994, 236, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jatana, N.; Ascher, D.B.; Pires, D.E.; Gokhale, R.S.; Thukral, L. Human LC3 and GABARAP subfamily members achieve functional specificity via specific structural modulations. Autophagy 2020, 16, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, S.A.; Gelfand, V.I. 18 kDa microtubule-associated protein: Identification as a new light chain (LC-3) of microtubule-associated protein 1 (MAP-1). FEBS Lett. 1987, 212, 145–148. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Bedford, F.K.; Brandon, N.J.; Moss, S.J.; Olsen, R.W. GABA A-receptor-associated protein links GABA A receptors and the cytoskeleton. Nature 1999, 397, 69–72. [Google Scholar] [CrossRef]
- Weidberg, H.; Shvets, E.; Shpilka, T.; Shimron, F.; Shinder, V.; Elazar, Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010, 29, 1792–1802. [Google Scholar] [CrossRef]
- Grunwald, D.S.; Otto, N.M.; Park, J.-M.; Song, D.; Kim, D.-H. GABARAPs and LC3s have opposite roles in regulating ULK1 for autophagy induction. Autophagy 2020, 16, 600–614. [Google Scholar] [CrossRef]
- Joachim, J.; Jefferies, H.B.; Razi, M.; Frith, D.; Snijders, A.P.; Chakravarty, P.; Judith, D.; Tooze, S.A. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol. Cell 2015, 60, 899–913. [Google Scholar] [CrossRef] [Green Version]
- Holdgaard, S.G.; Cianfanelli, V.; Pupo, E.; Lambrughi, M.; Lubas, M.; Nielsen, J.C.; Eibes, S.; Maiani, E.; Harder, L.M.; Wesch, N.; et al. Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat. Commun. 2019, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Joachim, J.; Razi, M.; Judith, D.; Wirth, M.; Calamita, E.; Encheva, V.; Dynlacht, B.D.; Snijders, A.P.; O’Reilly, N.; Jefferies, H.B.; et al. Centriolar satellites control GABARAP ubiquitination and GABARAP-mediated autophagy. Curr. Biol. 2017, 27, 2123–2136.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Muhlinen, N.; Akutsu, M.; Ravenhill, B.J.; Foeglein, Á.; Bloor, S.; Rutherford, T.J.; Freund, S.M.; Komander, D.; Randow, F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell 2012, 48, 329–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, R.; De la Concepcion, J.C.; Maqbool, A.; Kamoun, S.; Dagdas, Y.F. ATG8 expansion: A driver of selective autophagy diversification? Trends Plant Sci. 2017, 22, 204–214. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple functions of ATG8 family proteins in plant autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef] [PubMed]
- Zess, E.K.; Jensen, C.; Cruz-Mireles, N.; De la Concepcion, J.C.; Sklenar, J.; Stephani, M.; Imre, R.; Roitinger, E.; Hughes, R.; Belhaj, K. N-terminal β-strand underpins biochemical specialization of an ATG8 isoform. PLoS Biol. 2019, 17, e3000373. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Tabata, K.; Morita, E.; Kawasaki, M.; Kato, R.; Dobson, R.C.; Yoshimori, T.; Wakatsuki, S. Structural basis of the autophagy-related LC3/Atg13 LIR complex: Recognition and interaction mechanism. Structure 2014, 22, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.V.; Suzuki, H.; Marinković, M.; Lang, V.; Kato, R.; Kawasaki, M.; Buljubašić, M.; Šprung, M.; Rogova, N.; Wakatsuki, S.; et al. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Shrestha, B.K.; Rasmussen, M.S.; Abudu, Y.P.; Bruun, J.-A.; Larsen, K.B.; Alemu, E.A.; Sjøttem, E.; Lamark, T.; Johansen, T. NIMA-related kinase 9–mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. J. Biol. Chem. 2020, 295, 1240–1260. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, D.S.; Jariwala, J.S.; Anderson, E.; Mitra, K.; Meisenhelder, J.; Chang, J.T.; Ideker, T.; Hunter, T.; Nizet, V.; Dillin, A.; et al. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol. Cell 2015, 57, 55–68. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.V.; Suzuki, H.; Fiskin, E.; Wild, P.; Kniss, A.; Rozenknop, A.; Kato, R.; Kawasaki, M.; McEwan, D.G.; Löhr, F.; et al. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 2013, 454, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, D.A. Cation-π interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science 1996, 271, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Genau, H.M.; Huber, J.; Baschieri, F.; Akutsu, M.; Dötsch, V.; Farhan, H.; Rogov, V.; Behrends, C. CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol. Cell 2015, 57, 995–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirth, M.; Zhang, W.; Razi, M.; Nyoni, L.; Joshi, D.; O’Reilly, N.; Johansen, T.; Tooze, S.A.; Mouilleron, S. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherra, S.J., 3rd; Kulich, S.M.; Uechi, G.; Balasubramani, M.; Mountzouris, J.; Day, B.W.; Chu, C.T. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010, 190, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Bhaskara, R.M.; Lystad, A.H.; Gestal-Mato, U.; Covarrubias-Pinto, A.; Bonn, F.; Simonsen, A.; Hummer, G.; Dikic, I. TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 2020, 21, e48317. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, D.; Liu, W.; Peng, J.; Feng, J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem. Biophys. Res. Commun. 2010, 395, 471–476. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J.; et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–466. [Google Scholar] [CrossRef] [Green Version]
- Jia, R.; Bonifacino, J.S. Negative regulation of autophagy by UBA6-BIRC6–mediated ubiquitination of LC3. Elife 2019, 8, e50034. [Google Scholar] [CrossRef]
- Alemu, E.A.; Lamark, T.; Torgersen, K.M.; Birgisdottir, A.B.; Larsen, K.B.; Jain, A.; Olsvik, H.; Øvervatn, A.; Kirkin, V.; Johansen, T. ATG8 family proteins act as scaffolds for assembly of the ULK complex sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012, 287, 39275–39290. [Google Scholar] [CrossRef] [Green Version]
- Birgisdottir, Å.B.; Mouilleron, S.; Bhujabal, Z.; Wirth, M.; Sjøttem, E.; Evjen, G.; Zhang, W.; Lee, R.; O’Reilly, N.; Tooze, S.A.; et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 2019, 15, 1333–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozenknop, A.; Rogov, V.V.; Rogova, N.Y.; Löhr, F.; Güntert, P.; Dikic, I.; Dötsch, V. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 2011, 410, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Di Rita, A.; Peschiaroli, A.; Pasquale, D.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Ma, K.; Zhou, C.; Ding, P.; Zhu, Y.; Chen, Q.; Xia, B. Structural basis for the phosphorylation of FUNDC1 LIR as a molecular switch of mitophagy. Autophagy 2016, 12, 2363–2373. [Google Scholar] [CrossRef] [Green Version]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Massen, S.; Terenzio, M.; Lang, V.; Chen-Lindner, S.; Eils, R.; Novak, I.; Dikic, I.; Hamacher-Brady, A.; Brady, N.R. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 2013, 288, 1099–1113. [Google Scholar] [CrossRef] [Green Version]
- Sano, T.; Cantor, C.R. Cooperative biotin binding by streptavidin. Electrophoretic behavior and subunit association of streptavidin in the presence of 6 M urea. J. Biol. Chem. 1990, 265, 3369–3373. [Google Scholar]
- Stubenrauch, K.; Wessels, U.; Essig, U.; Kowalewsky, F.; Vogel, R.; Heinrich, J. Characterization of murine anti-human Fab antibodies for use in an immunoassay for generic quantification of human Fab fragments in non-human serum samples including cynomolgus monkey samples. J. Pharm. Biomed. Anal. 2013, 72, 208–215. [Google Scholar] [CrossRef]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-binding domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef]
- Li, J.; Zhu, R.; Chen, K.; Zheng, H.; Zhao, H.; Yuan, C.; Zhang, H.; Wang, C.; Zhang, M. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat. Chem. Biol. 2018, 14, 778–787. [Google Scholar] [CrossRef]
- Xie, Q.; Tzfadia, O.; Levy, M.; Weithorn, E.; Peled-Zehavi, H.; Van Parys, T.; Van de Peer, Y.; Galili, G. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 2016, 12, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Habisov, S.; Huber, J.; Ichimura, Y.; Akutsu, M.; Rogova, N.; Loehr, F.; McEwan, D.G.; Johansen, T.; Dikic, I.; Doetsch, V.; et al. Structural and functional analysis of a novel interaction motif within UFM1-activating enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and ufmylation. J. Biol. Chem. 2016, 291, 9025–9041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumbarello, D.A.; Manna, P.T.; Allen, M.; Bycroft, M.; Arden, S.D.; Kendrick-Jones, J.; Buss, F. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of salmonella typhimurium by autophagy. PLoS Pathog 2015, 11, e1005174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufmann, A.; Beier, V.; Franquelim, H.G.; Wollert, T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell 2014, 156, 469–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, N.N.; Satoo, K.; Fujioka, Y.; Kumeta, H.; Ogura, K.; Nakatogawa, H.; Ohsumi, Y.; Inagaki, F. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 2011, 44, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, Y.; Gong, Y.; Li, F.; Guo, Y.; Hu, S.; Liu, J.; Pan, L. Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins. Autophagy 2016, 12, 1330–1339. [Google Scholar] [CrossRef] [Green Version]
- Khaminets, A.; Heinrich, T.; Mari, M.; Grumati, P.; Huebner, A.K.; Akutsu, M.; Liebmann, L.; Stolz, A.; Nietzsche, S.; Koch, N.; et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015, 522, 354–358. [Google Scholar] [CrossRef]
- Olsvik, H.L.; Lamark, T.; Takagi, K.; Larsen, K.B.; Evjen, G.; Øvervatn, A.; Mizushima, T.; Johansen, T. FYCO1 contains a C-terminally extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy. J. Biol. Chem. 2015, 290, 29361–29374. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, S.; Tomita, T.; Shimizu, T.; Ohto, U. The crystal structure of mouse LC3B in complex with the FYCO1 LIR reveals the importance of the flanking region of the LIR motif. Acta Cryst. F 2017, 73, 130–137. [Google Scholar] [CrossRef]
- Mochida, K.; Yamasaki, A.; Matoba, K.; Kirisako, H.; Noda, N.N.; Nakatogawa, H. Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Nelson, A.D.; Caballero-Florán, R.N.; Díaz, J.R.; Hull, J.; Yuan, Y.; Li, J.; Chen, K.; Walder, K.; Lopez-Santiago, L.; Bennett, V.; et al. Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP. Mol. Psychiatry 2018, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Kim, L.; Kim, B.-W.; Kim, J.H.; Roh, K.-H.; Choi, E.-J.; Song, H.K. A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ. Biochem. Biophys. Res. Commun. 2017, 490, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Lystad, A.H.; Ichimura, Y.; Takagi, K.; Yang, Y.; Pankiv, S.; Kanegae, Y.; Kageyama, S.; Suzuki, M.; Saito, I.; Mizushima, T.; et al. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC 3B-positive structures. EMBO Rep. 2014, 15, 557–565. [Google Scholar] [CrossRef] [Green Version]
- Keown, J.R.; Black, M.M.; Ferron, A.; Yap, M.; Barnett, M.J.; Pearce, F.G.; Stoye, J.P.; Goldstone, D.C. A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5α and mammalian autophagy-related ATG8 proteins. J. Biol. Chem. 2018, 293, 18378–18386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, P.; Schwarten, M.; Schneider, L.; Boeske, A.; Henke, N.; Lisak, D.; Weber, S.; Mohrlüder, J.; Stoldt, M.; Strodel, B.; et al. Interaction of Bcl-2 with the Autophagy-related GABAA Receptor-associated Protein (GABARAP): Biophysical characterization and functional implications. J. Biol. Chem. 2013, 288, 37204–37215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Real, E.; Rodrigues, L.; Cabal, G.G.; Enguita, F.J.; Mancio-Silva, L.; Mello-Vieira, J.; Beatty, W.; Vera, I.M.; Zuzarte-Luís, V.; Figueira, T.N.; et al. Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat. Microbiol. 2018, 3, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Weiergräber, O.H.; Stangler, T.; Thielmann, Y.; Mohrlüder, J.; Wiesehan, K.; Willbold, D. Ligand binding mode of GABAA receptor-associated protein. J. Mol. Biol. 2008, 381, 1320–1331. [Google Scholar] [CrossRef]
- Stolz, A.; Putyrski, M.; Kutle, I.; Huber, J.; Wang, C.; Major, V.; Sidhu, S.S.; Youle, R.J.; Rogov, V.V.; Dötsch, V.; et al. Fluorescence-based ATG 8 sensors monitor localization and function of LC 3/GABARAP proteins. EMBO J. 2017, 36, 549–564. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.; Schillinger, O.; Schwarten, M.; Lecher, J.; Hartmann, R.; Stoldt, M.; Mohrlüder, J.; Olubiyi, O.; Strodel, B.; Willbold, D. Conformational polymorphism in autophagy-related protein GATE-16. Biochemistry 2015, 54, 5469–5479. [Google Scholar] [CrossRef]
- Shvets, E.; Abada, A.; Weidberg, H.; Elazar, Z. Dissecting the involvement of LC3B and GATE-16 in p62 recruitment into autophagosomes. Autophagy 2011, 7, 683–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Li, J.-G.; Chen, Y.; Huang, P.; Wang, Y.; Liu-Chen, L.-Y. GEC1 interacts with the κ opioid receptor and enhances expression of the receptor. J. Biol. Chem. 2006, 281, 7983–7993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kneussel, M.; Haverkamp, S.; Fuhrmann, J.C.; Wang, H.; Wässle, H.; Olsen, R.W.; Betz, H. The γ-aminobutyric acid type A receptor (GABAAR)-associated protein GABARAP interacts with gephyrin but is not involved in receptor anchoring at the synapse. Proc. Natl. Acad. Sci. USA 2000, 97, 8594–8599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, O.; Akpinar, H.A.; Rogov, V.V.; Kirkin, V. Selective autophagy receptors in neuronal health and disease. J. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C.; Wang, Z.; Zhu, C.; Li, J.; Sha, T.; Ma, L.; Gao, C.; Yang, Y.; Sun, Y.; et al. Allele-selective lowering of mutant HTT protein by HTT–LC3 linker compounds. Nature 2019, 575, 203–209. [Google Scholar] [CrossRef]
- Hu, D.; Wu, J.; Zhang, R.; Chen, L.; Chen, Z.; Wang, X.; Xu, L.; Xiao, J.; Hu, F.; Wu, C. Autophagy-targeted vaccine of LC3–LpqH DNA and its protective immunity in a murine model of tuberculosis. Vaccine 2014, 32, 2308–2314. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Venida, A.; Yano, J.; Biancur, D.E.; Kakiuchi, M.; Gupta, S.; Sohn, A.S.; Mukhopadhyay, S.; Lin, E.Y.; Parker, S.J.; et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020, 581, 100–105. [Google Scholar] [CrossRef]
- Park, S.-W.; Jeon, P.; Jun, Y.-W.; Park, J.-H.; Lee, S.-H.; Lee, S.; Lee, J.-A.; Jang, D.-J. Monitoring LC3-or GABARAP-positive autophagic membranes using modified RavZ-based probes. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]





| Protein | LC3A | LC3B | LC3C | GABARAP | GABARAPL1 | GABARAPL2 | Method | Ref. |
|---|---|---|---|---|---|---|---|---|
| AnkG | 0.55 | 0.34 | 2.39 | 2.6 × 10−3 | 3.7 × 10−3 | 40 × 10−3 | ITC | [100] |
| AnkB | 3.7 × 10−3 | 4.2 × 10−3 | 10.5 × 10−3 | 0.27 × 10−3 | 0.29 × 10−3 | 0.21 × 10−3 | ITC | [100] |
| PLEKHM1 | 4.22 | 6.33 | 3.45 | 0.55 | 0.77 | 0.93 | ITC | [34] |
| PCM1 | 292 | 982 | 17.9 | 2.0 | 1.6 | 14.4 | BLI | [84] |
| ULK1 | 5.9 | 48.2 | 2.5 | 50 × 10−3 | 48 × 10−3 | 0.53 | BLI | [84] |
| ATG13 | 4.1 | 9.6 | 0.48 | 0.59 | 0.53 | 3.1 | BLI | [84] |
| FIP200 | 281 | 1206 | 63.3 | 5.6 | 7.0 | 86.4 | BLI | [84] |
| p62/SQSTM1 | 2.0 | 4.5 | 2.7 | 0.9 | 0.6 | 5.2 | BLI | [84] |
| Ambra1 | >100 | >100 | >100 | 39 | >50 | >100 | ITC | [93] |
| pS1014Ambra1 | (50) | (50) | (>100) | (21) | (25) | (>100) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wesch, N.; Kirkin, V.; Rogov, V.V. Atg8-Family Proteins—Structural Features and Molecular Interactions in Autophagy and Beyond. Cells 2020, 9, 2008. https://doi.org/10.3390/cells9092008
Wesch N, Kirkin V, Rogov VV. Atg8-Family Proteins—Structural Features and Molecular Interactions in Autophagy and Beyond. Cells. 2020; 9(9):2008. https://doi.org/10.3390/cells9092008
Chicago/Turabian StyleWesch, Nicole, Vladimir Kirkin, and Vladimir V. Rogov. 2020. "Atg8-Family Proteins—Structural Features and Molecular Interactions in Autophagy and Beyond" Cells 9, no. 9: 2008. https://doi.org/10.3390/cells9092008
APA StyleWesch, N., Kirkin, V., & Rogov, V. V. (2020). Atg8-Family Proteins—Structural Features and Molecular Interactions in Autophagy and Beyond. Cells, 9(9), 2008. https://doi.org/10.3390/cells9092008