GWAS Discovery of Candidate Genes for Yield-Related Traits in Peanut and Support from Earlier QTL Mapping Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Statistics
2.3. Genotyping by Sequencing of Peanut Cultivars
2.4. Population Genetic Analysis
2.5. Genome-Wide Association Study Analysis
2.6. Candidate Gene Identification
3. Results
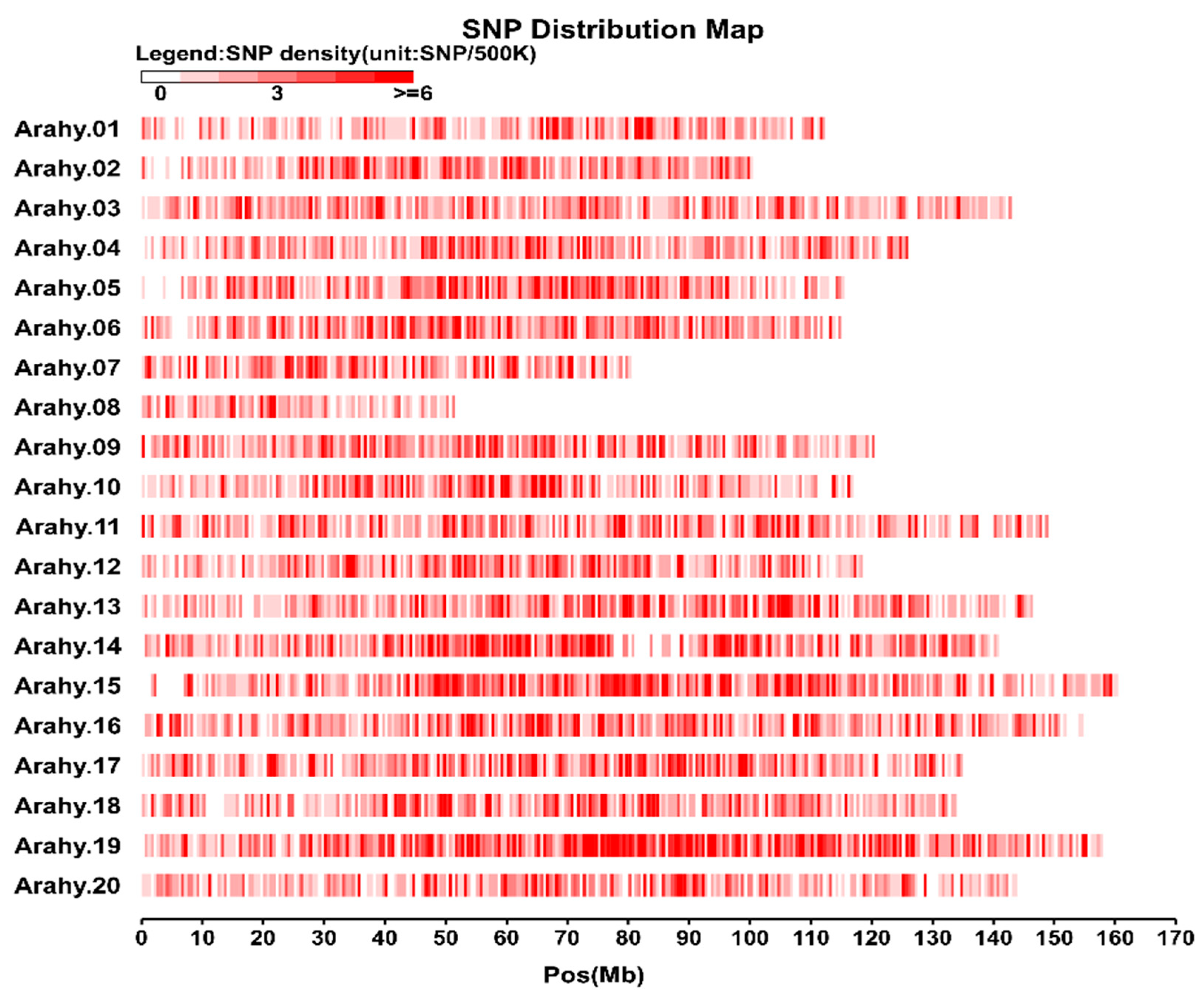
3.1. Characterisation and Distribution of SNPs in The Peanut Genome
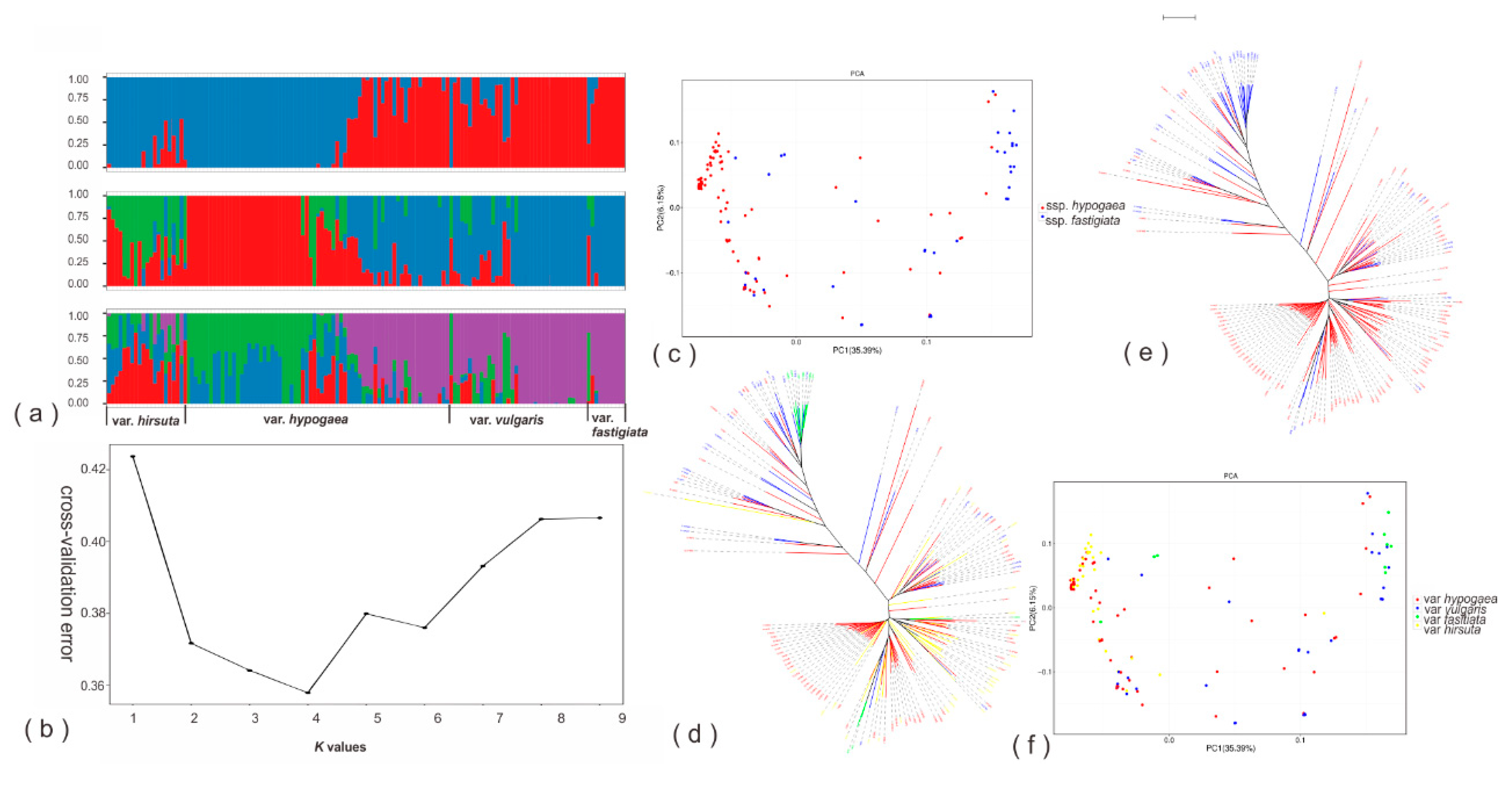
3.2. Genetic Diversity, Population Structure, and Genetic Diversity
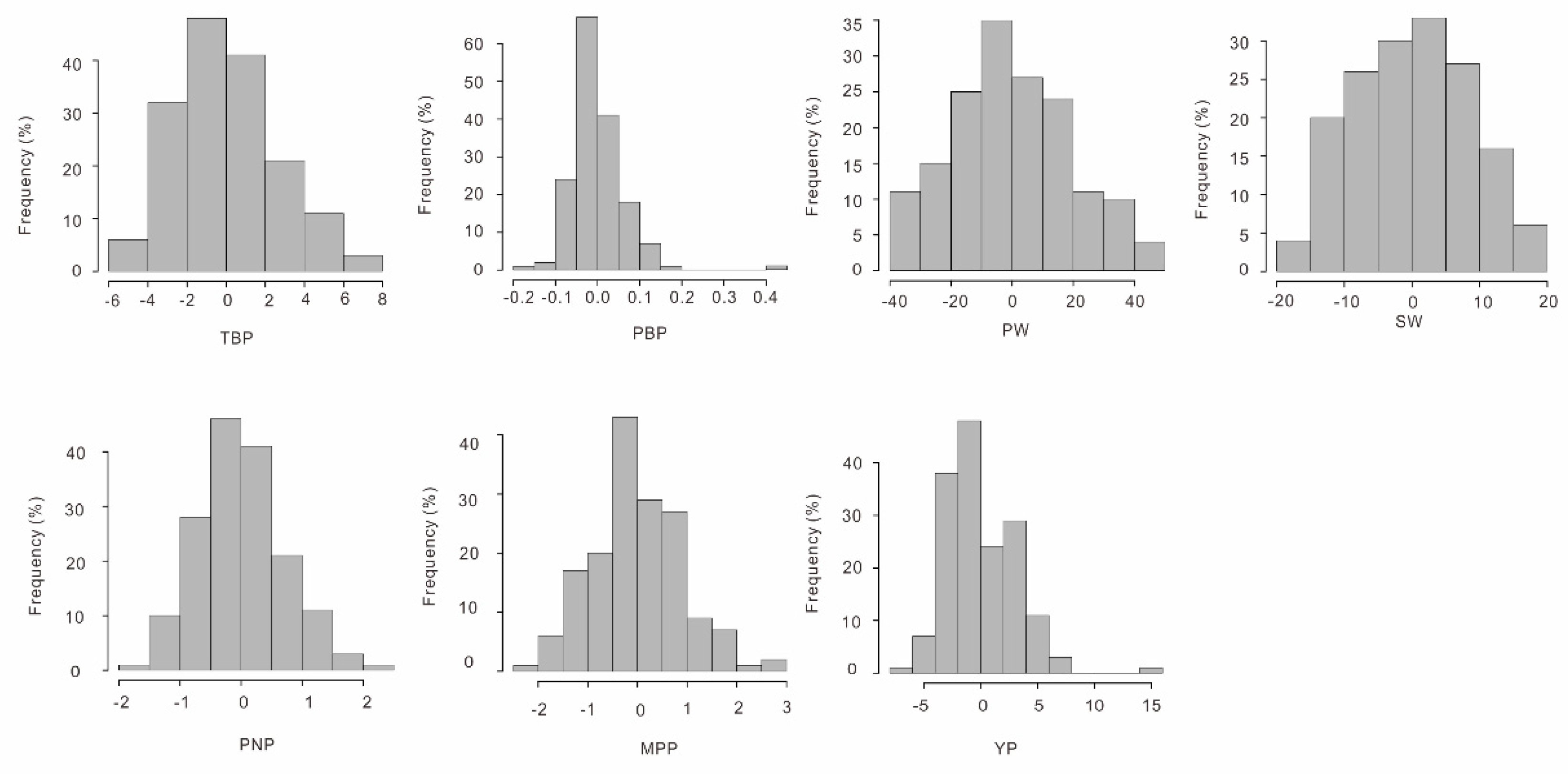
3.3. Phenotypic Correlation and Heritability for Different Traits
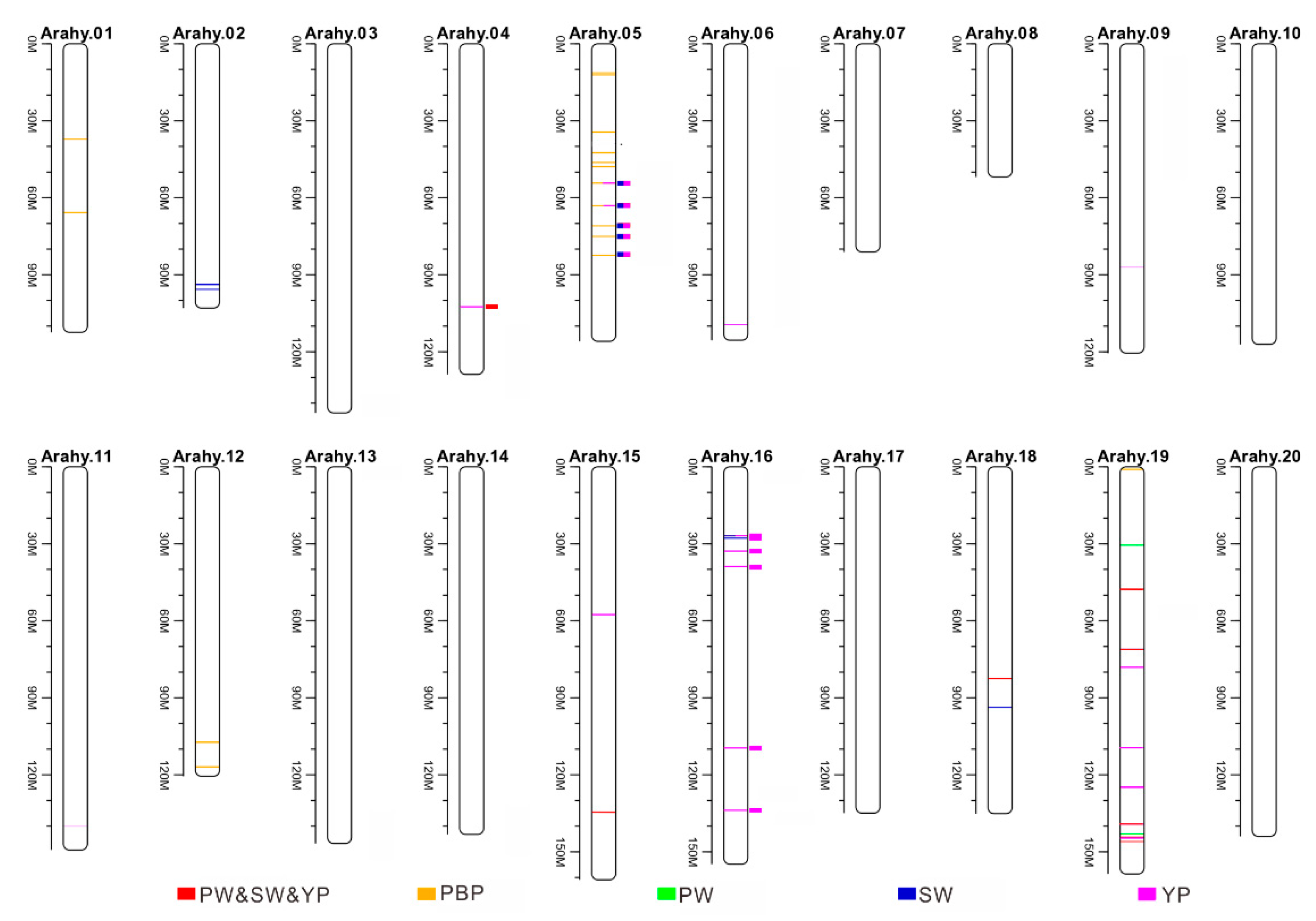
3.4. Genome-Wide Association Studies in Peanut
3.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
3.6. Literature Survey of Previous Identified QTLs
4. Discussion
4.1. Genome-Wide SNP Discovery
 T, A
T, A G) is therefore conformationally more favorable than that generated by transversions (A
G) is therefore conformationally more favorable than that generated by transversions (A C, C
C, C G, A
G, A T, G
T, G T) [51]. Moreover, transitions give rise to less amino acid replacements and tend to conserve the chemical property of an amino acid if it does bring on amino acid replacements.
T) [51]. Moreover, transitions give rise to less amino acid replacements and tend to conserve the chemical property of an amino acid if it does bring on amino acid replacements.4.2. Population Structure
4.3. Yield-Related Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Culbreath, A. Why preserve and evaluate genetic resources in peanut? International Conference of the peanut research community on advances in Arachis through Genomics & Biotechnology; USDA: Washington, DC, USA, 2013. [Google Scholar]
- Yu, S.L. Peanut Genetics and Breeding in China; Shanghai Science and Technology Press: Shanghai, China, 2011; pp. 11–13. [Google Scholar]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Mohraan, S.M.; Gaur, P.M.; Gangarao, N.V.P.R.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Kover, P.X.; Valdar, W.; Trakalo, J.; Scarcelli, N.; Ehrenreich, I.M.; Putugganan, M.D.; Durrant, C.; Mon, R. A multiparent advanced generation inter-cross to fine map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000551. [Google Scholar] [CrossRef] [PubMed]
- Khedikar, Y.; Pandey, M.K.; Sujay, V.; Singh, S.; Nayak, S.N.; Klein-Gebbinck, H.W.; Sarvamangala, C.; Mukri, G.; Garg, V.; Upadhyaya, H.D.; et al. Identification of main effect and epistatic quantitative trait loci for morphological and yield-related traits in peanut (Arachis hypogaea L.). Mol. Breed. 2018, 38, 7. [Google Scholar] [CrossRef]
- Huang, L.; He, H.; Chen, W.; Ren, X.; Chen, Y.; Zhou, X.; Xia, Y.; Wang, X.; Jiang, X.; Liao, B.S.; et al. Quantitative trait locus analysis of agronomic and quality-related traits in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2015, 128, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Hake, A.A.; Kenta, S.; Arati, Y.; Sukruth, M.; Malagouda, P.; Nayak, S.N.; Lingaraju, S.; Patil, P.V.; Nadaf, H.L.; Gowda, M.V.C.; et al. Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L.). PLoS ONE 2017, 12, e0186113. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.Y.; Ren, X.P.; Li, Z.D.; Xu, Z.J.; Li, X.P.; Huang, L.; Zhou, X.J.; Chen, Y.N.; Chen, W.G.; Lei, Y.; et al. Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.). BMC Genom. 2017, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.G.; Zhang, J.H.; He, X.Y.; Wang, Y.; Ma, X.L.; Yin, D.M. Genome-wide association study of major agronomic traits related to domestication in Peanut. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.Y.; Abdukarimov, A. Application of association mapping to understanding the genetic diversity of plant germplasm resources. Int. J. Plant Genom. 2008, 2008, 1–18. [Google Scholar] [CrossRef]
- Pandey, M.K.; Upadhyaya, H.D.; Rathore, A.; Vadez, V.; Sheshshaye, M.S.; Sriswathi, M.; Govil, M.; Kumar, A.; Gowda, M.V.C.; Shivali, S.; et al. Genome-wide association studies for 50 agronomic traits in peanut using the reference set comprising 300 genotypes from 48countries of semi-arid tropics of the world. PLoS ONE 2014, 9, e105228. [Google Scholar] [CrossRef]
- Yan, M.; Zhang, X.Y.; Han, S.Y.; Huang, B.; Dong, W.; Liu, H.; Sun, Z.Q.; Zhang, Z.X.; Tang, F.S. Genome-wide association study of agronomic and yield traits in a worldwide collection of peanut (Arachis hypogaea L.) Germplasm. Chin. Bull. Bot. 2015, 50, 460–472. (In Chinese) [Google Scholar]
- Zhao, J.J.; Huang, L.; Ren, X.P.; Pandey, M.K.; Wu, B.; Chen, Y.; Zhou, X.; Chen, W.; Xia, Y.; Li, Z.; et al. Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) Using Single-Locus Simple Sequence Repeat Markers. Front. Plant Sci. 2017, 11, 2105. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Agarwal, G.; Kale, S.M.; Clevenger, J.; Nayak, S.N.; Sriswathi, M.; Chitikineni, A.; Chavarro, C.; Chen, X.; Upadhyaya, H.D. Development and evaluation of a high density genotyping Axiom arachis array with 58K SNPs for accelerating genetics and breeding in groundnut. Sci. Rep. 2017, 7, 40577. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, Z.Q.; Fang, Y.J.; Qi, F.Y.; Liu, H.; Miao, L.J.; Du, P.; Shi, L.; Gao, W.; Han, S.Y.; et al. Genetic diversity, population structure, and botanical variety of 320 global peanut accessions revealed through tunable genotyping-by-sequencing. Sci. Rep. 2018, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Kump, K.L.; Bradbury, P.J.; Wisser, R.J.; Buckler, E.S.; Belcher, A.R.; Oropeza-Rosas, M.A.; Zwonitzer, J.C.; Kresovich, S.; McMullen, M.K.; Ware, D.; et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 2011, 43, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 13, 211–221. [Google Scholar] [CrossRef]
- Fang, C.; Ma, Y.; Wu, S.; Liu, Z.; Wang, Z.; Yang, R.; Hu, G.H.; Zhou, Z.K.; Yu, H.; Zhang, M.; et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017, 18, 161. [Google Scholar] [CrossRef]
- Su, J.J.; Pang, C.Y.; Wei, H.L.; Li, L.L.; Liang, B.; Wang, C.X.; Song, M.Z.; Wang, H.T.; Zhao, S.Q.; Jia, X.Y.; et al. Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton. BMC Genom. 2016, 17, 687. [Google Scholar] [CrossRef]
- Su, J.; Fan, S.; Li, L.; Wei, H.; Wang, C.; Wang, H.; Song, M.; Zhang, C.; Gu, L.; Zhao, S.; et al. Detection of favorable QTL alleles and candidate genes for lint percentage by GWAS in Chinese upland cotton. Front. Plant Sci. 2016, 7, 1576. [Google Scholar] [CrossRef]
- Ariani, A.; Teran, J.C.B.M.Y.; Gepts, P. Genome-wide identification of SNPs and copy number variation in common bean (Phaseolus vulgaris L.) using genotyping-by-sequencing (GBS). Mol. Breed. 2017, 36, 87. [Google Scholar] [CrossRef]
- Zhuang, W.J.; Chen, H.; Yang, M.; Wang, J.P.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.S.; Zhang, X.T.; Tang, R.H.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.H.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Construction of core collections and genetic diversity analysis of Arachis hypogaea L. cultivars in China. Master’s Thesis, ZhongKai college of Agriculture and Engineering, Guangzhou, China, 2013; pp. 1–36. [Google Scholar]
- Jiang, H.F.; Duan, N.; Ren, X.P. Descriptors and Data Standard for Peanut (Arachis spp.); China Agriculture Press: Beijing, China, 2006. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; ISBN 3-900051-07-0. [Google Scholar]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.J.; Yan, C.X.; Zhao, X.B.; Shan, S.H. The screening of ddGBS restriction enzyme combinations in peanut. J. Peanut Sci. 2017, 46, 48–51. (In Chinese) [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013, 43, 11–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.Y.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.65; University of Washington: Seattle, WA, USA, 2005. [Google Scholar]
- Lee, S.H.; Yang, J.; Goddard, M.E.; Visscher, P.M.; Wray, N.R. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 2012, 28, 2540–2542. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyses spatial genetic structure at the individual or population levels. Mol. Ecol. Resour. 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. Tassel: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.; Casstevens, T.; Lu, F. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, 90346. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Stalker, H.T. Utilizing wild species for peanut improvement. Crop. Sci. 2017, 57, 1102–1120. [Google Scholar] [CrossRef]
- Stalker, H.T.; Simpson, C.E. Germplasm resources in peanut (Arachis hypogaea). In Advances in Peanut Science; Pattee, H.E., Stalker, H.T., Eds.; American Peanut Research and Education Society: Stillwater, OK, USA, 1995; pp. 14–53. [Google Scholar]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An efficient method of large-scale De Novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef] [PubMed]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016. [Google Scholar] [CrossRef]
- Morton, B.R. Neighboring base composition and transversion ⁄transition bias in a comparison of rice and maize chloroplast noncoding regions. Proc. Natl. Acad. Sci. USA 1995, 92, 9717–9721. [Google Scholar] [CrossRef]
- Batley, J.; Barker, G.; O’Sullivan, H.; Edwards, K.J.; Edwards, D. Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 2003, 132, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Uitdewilligen, J.G.; Wolters, A.M.; D’Hoop, B.B.; Borm, T.J.; Visser, R.G.; van Eck, H.J. A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS ONE 2013, 8, e62355. [Google Scholar] [CrossRef] [PubMed]
- Taranto, F.; D’Agostino, N.; Greco, B.; Cardi, T.; Tripodi, P. Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genom. 2016, 17, 943. [Google Scholar] [CrossRef] [PubMed]
- Wakeley, J. The excess of transitions among nucleotide substitutions: New methods of estimating transition bias underscore its significance. Trends Ecol. Evol. 1996, 11, 158–162. [Google Scholar] [CrossRef]
- Krapovickas, A.; Gregory, W.C. Taxonomia del genero Arachis (Leguminosae). Bonplandia 1994, 8, 1–186. [Google Scholar] [CrossRef]
- Krapovickas, A.; Gregory, W.C.; Williams, D.E.; Simpson, C.E. Taxonomy of the genus Arachis (Leguminosae). Bonplandia 2007, 16, 7–205. [Google Scholar] [CrossRef]
- Li, S.L. The pedigree analysis of Peanut varieties. Peanut Sci. Technol. 2000, 4. [Google Scholar] [CrossRef]
- Sun, Z.Q.; Qi, F.Y.; Zheng, Z.; Dong, W.Z.; Huang, B.Y.; Zhang, J. Analysis of parental relationship of 106 peanut cultivars released in Henan province. Chin. J. Oil Crop. Sci. 2017, 39, 754–762. [Google Scholar] [CrossRef]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shetn, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in price increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]

| Traits | Abbr. | Maximum | Minimum | Median | Average | Variance | SD | CV (%) | H2 |
|---|---|---|---|---|---|---|---|---|---|
| Total Branching Number | TBN | 31.27 | 9.26 | 18.34 | 18.80 | 19.81 | 6.28 | 33.00 | 0.54 |
| Pod Branching Number | PBP | 16.16 | 4.84 | 7.93 | 8.07 | 1.57 | 1.25 | 16.00 | 0.04 |
| Pods Number Per Plant | PNP | 38.97 | 15.58 | 24.16 | 24.46 | 17.19 | 4.15 | 17.00 | 0.14 |
| Mature Pods Number Per Plant | MPP | 31.89 | 10.34 | 19.02 | 19.38 | 14.60 | 3.82 | 20.00 | 0.19 |
| 100-Pod Weight | PW | 235.16 | 100.73 | 159.24 | 160.55 | 859.34 | 29.31 | 18.00 | 0.63 |
| 100-Seed Weigh | SW | 84.54 | 35.93 | 59.55 | 59.20 | 125.03 | 11.18 | 7.00 | 0.72 |
| Yield Per Plant | YP | 60.86 | 12.12 | 25.34 | 26.95 | 47.54 | 6.90 | 26.00 | 0.39 |
| The Present Study | Earlier Studies | |||||
|---|---|---|---|---|---|---|
| Peak SNP | Position (± 100 kb) | Associated Traits | Annotated Gene List | Co-localized QTLsa | Studied Traits | Ref. |
| rs2123 | Arahy.04: 102215951 | YP | arahy.Q2K3EA; arahy.JMQ6FC | Arahy.04: 94872059-124685617 | PW;SW;YP | [1] |
| rs2500 | Arahy.05: 54266788 | PBP; YP | arahy.T4811Q; arahy.71GWZM | Arahy.05: 47955696-84171615; | YP; SW | [23] |
| rs2560 | Arahy.05: 63009987 | PBP; YP | arahy.A6MV6T | Arahy.05: 47955696-84171615; 6445048-6913313 | YP; SW | [1] |
| rs2639 | Arahy.05: 70800888 | PBP | arahy.1NMA8N; arahy.J02DSU | Arahy.05: 47955696-84171615; | YP; SW | [23] |
| rs2675 | Arahy.05: 75058945 | PBP | arahy.7FT54I; arahy.D7MP66; arahy.V7ESBJ | Arahy.05: 47955696-84171615; | YP; SW | [1] |
| rs2725 | Arahy.05: 82267261 | PBP | arahy.LH3A9J; arahy.NM6FQB; arahy.AVNN4W | Arahy.05: 47955696-84171615; | YP; SW | [23] |
| rs9379 | Arahy.16: 26887159 | SW, YP | arahy.7B5I5W | Arahy.16: 16836027-41668600 | YP | [23] |
| rs9380 | Arahy.16: 26954799 | SW | arahy.KBV6L0; arahy.Y4GE00; arahy.A3WBL0; arahy.H97NC3 | Arahy.16: 16836027-41668600 | YP | [23] |
| rs9411 | Arahy.16: 32888295 | YP | arahy.RI9HIF; arahy.HCD6SP; arahy.9BIX4A; arahy.N88R54 | Arahy.16: 16836027-41668600 | YP | [23] |
| rs9432 | Arahy.16: 38878752 | YP | arahy.SDZN59; arahy.KW19LS; arahy.FC0UQU; arahy.PY3JPZ; arahy.05RLXM; arahy.K0A92D | Arahy.16: 16836027-41668600 | YP | [23] |
| rs9906 | Arahy.16: 109497037 | YP | arahy.60591L | Arahy.16: 16836027-141668600 | YP | [23] |
| rs10035 | Arahy.16: 133839995 | YP | arahy.84A022; arahy.V06H0G; arahy.C96CVE; arahy.7GY2ZZ; arahy.A4HJXM; arahy.QZI7QE; arahy.AA3MKH | Arahy.16:16836027-141668601 | YP | [23] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Yan, C.; Li, Y.; Li, C.; Zhao, X.; Yuan, C.; Sun, Q.; Shan, S. GWAS Discovery of Candidate Genes for Yield-Related Traits in Peanut and Support from Earlier QTL Mapping Studies. Genes 2019, 10, 803. https://doi.org/10.3390/genes10100803
Wang J, Yan C, Li Y, Li C, Zhao X, Yuan C, Sun Q, Shan S. GWAS Discovery of Candidate Genes for Yield-Related Traits in Peanut and Support from Earlier QTL Mapping Studies. Genes. 2019; 10(10):803. https://doi.org/10.3390/genes10100803
Chicago/Turabian StyleWang, Juan, Caixia Yan, Yuan Li, Chunjuan Li, Xiaobo Zhao, Cuiling Yuan, Quanxi Sun, and Shihua Shan. 2019. "GWAS Discovery of Candidate Genes for Yield-Related Traits in Peanut and Support from Earlier QTL Mapping Studies" Genes 10, no. 10: 803. https://doi.org/10.3390/genes10100803
APA StyleWang, J., Yan, C., Li, Y., Li, C., Zhao, X., Yuan, C., Sun, Q., & Shan, S. (2019). GWAS Discovery of Candidate Genes for Yield-Related Traits in Peanut and Support from Earlier QTL Mapping Studies. Genes, 10(10), 803. https://doi.org/10.3390/genes10100803





