Roles of Topoisomerases in Heterochromatin, Aging, and Diseases
Abstract
1. Introduction
2. Importance of Heterochromatin
2.1. Heterochromatin Is Critical for Transcriptional Silencing of Transposons
2.2. Loss of Heterochromatin May Be a Cause of Aging and Premature Aging Syndromes
2.3. Heterochromatin Loss May Increase Cancer Risks
2.4. Heterochromatin and Neurological Disorders
3. Multiple Topoisomerases Function in Heterochromatin
3.1. Top1
3.2. Top2
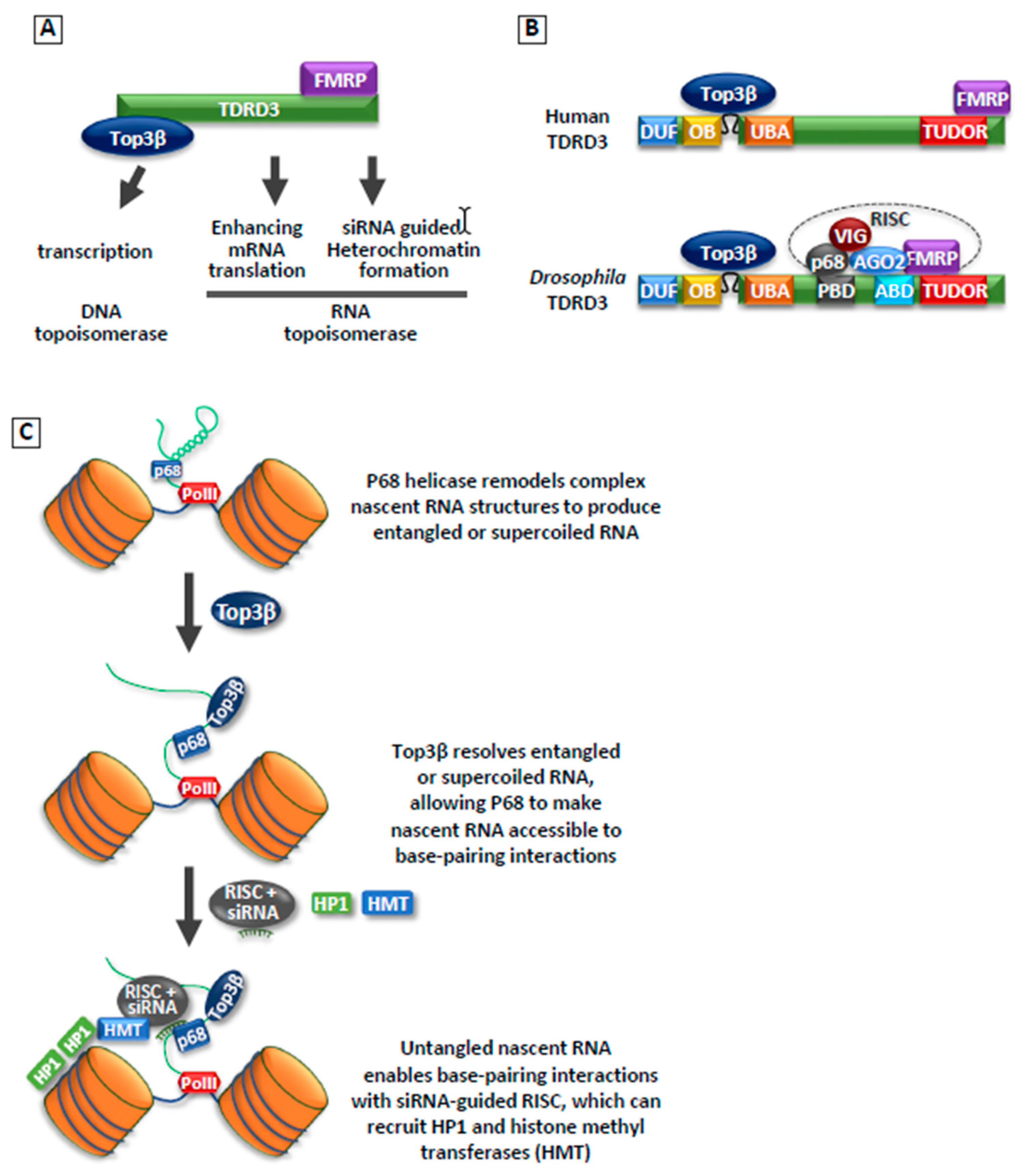
3.3. Top3β
3.3.1. Top3β Is a Dual-Activity Topoisomerase
3.3.2. Top3β Is Required for Heterochromatin Formation and Silencing of Transposons
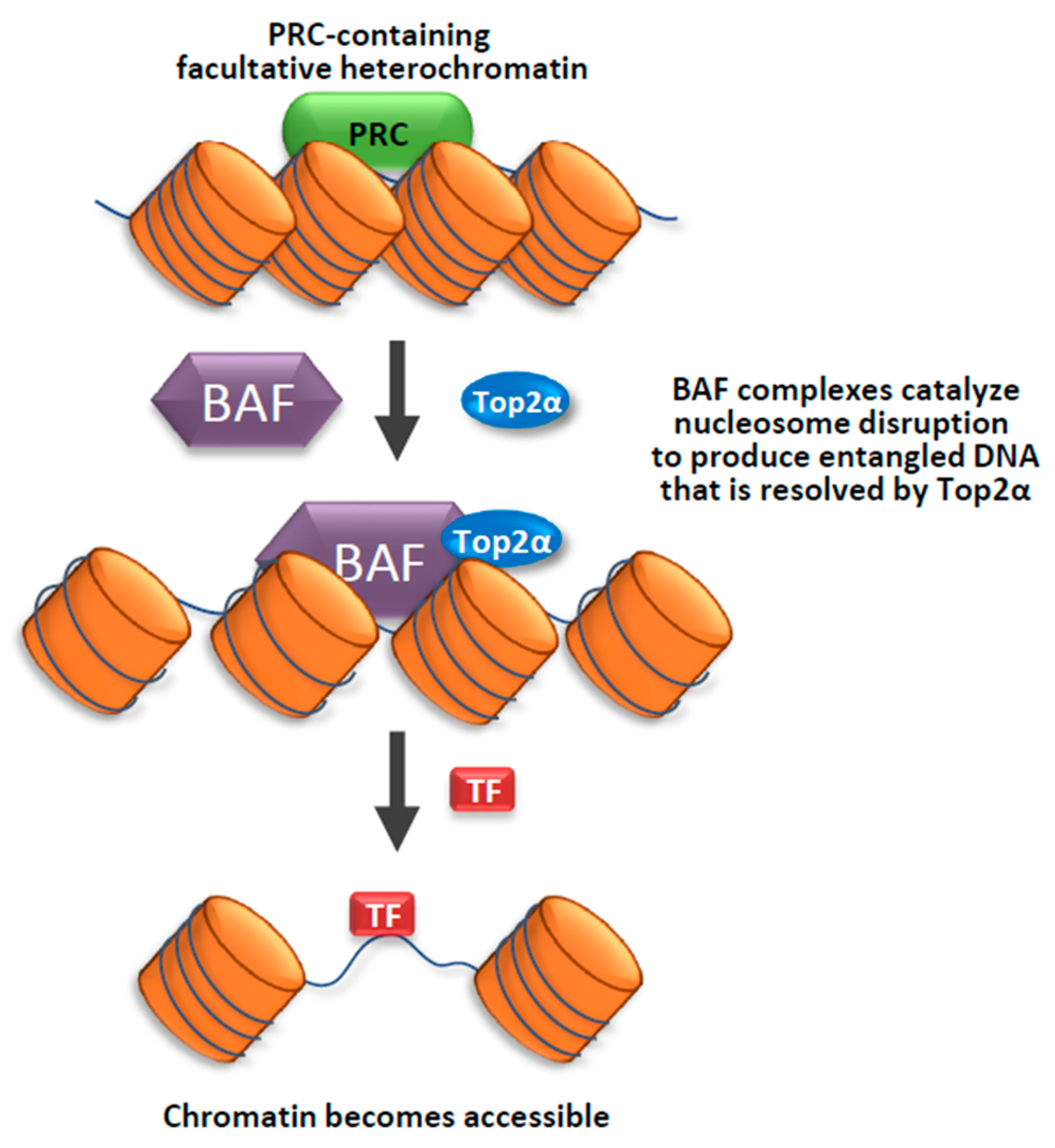
4. Importance of Topoisomerase Function in Heterochromatin
5. Conclusive Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Chen, A.D.; Gettel, N.J.; Hsieh, T.S. Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev. Biol. 2000, 222, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Morham, S.G.; Kluckman, K.D.; Voulomanos, N.; Smithies, O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell Biol. 1996, 16, 6804–6809. [Google Scholar] [CrossRef] [PubMed]
- Hohl, A.M.; Thompson, M.; Soshnev, A.A.; Wu, J.; Morris, J.; Hsieh, T.S.; Wu, C.T.; Geyer, P.K. Restoration of topoisomerase 2 function by complementation of defective monomers in Drosophila. Genetics 2012, 192, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.Y.; Moens, P.B.; Wang, J.C. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III β. Proc. Natl. Acad. Sci. USA 2003, 100, 2526–2531. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.Y.; Wang, J.C. Mice lacking DNA topoisomerase IIIβ develop to maturity but show a reduced mean lifespan. Proc. Natl. Acad. Sci. USA 2001, 98, 5717–5721. [Google Scholar] [CrossRef] [PubMed]
- Kwan, K.Y.; Greenwald, R.J.; Mohanty, S.; Sharpe, A.H.; Shaw, A.C.; Wang, J.C. Development of autoimmunity in mice lacking DNA topoisomerase 3β. Proc. Natl. Acad. Sci. USA 2007, 104, 9242–9247. [Google Scholar] [CrossRef]
- Xu, D.; Shen, W.; Guo, R.; Xue, Y.; Peng, W.; Sima, J.; Yang, J.; Sharov, A.; Srikantan, S.; Yang, J.; et al. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013, 16, 1238–1247. [Google Scholar] [CrossRef]
- Li, M.; Liu, Y. Topoisomerase I in Human Disease Pathogenesis and Treatments. Genomics Proteomics Bioinform. 2016, 14, 166–171. [Google Scholar] [CrossRef]
- Lam, C.W.; Yeung, W.L.; Law, C.Y. Global developmental delay and intellectual disability associated with a de novo TOP2B mutation. Clin Chim Acta 2017, 469, 63–68. [Google Scholar] [CrossRef]
- Stoll, G.; Pietilainen, O.P.; Linder, B.; Suvisaari, J.; Brosi, C.; Hennah, W.; Leppa, V.; Torniainen, M.; Ripatti, S.; Ala-Mello, S.; et al. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013, 16, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013, 8, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Durand-Dubief, M.; Persson, J.; Norman, U.; Hartsuiker, E.; Ekwall, K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010, 29, 2126–2134. [Google Scholar] [CrossRef]
- Sperling, A.S.; Jeong, K.S.; Kitada, T.; Grunstein, M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 12693–12698. [Google Scholar] [CrossRef]
- Baranello, L.; Wojtowicz, D.; Cui, K.; Devaiah, B.N.; Chung, H.J.; Chan-Salis, K.Y.; Guha, R.; Wilson, K.; Zhang, X.; Zhang, H.; et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell 2016, 165, 357–371. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Burger, L.; Nikoletopoulou, V.; Deogracias, R.; Thakurela, S.; Wirbelauer, C.; Kaut, J.; Terranova, R.; Hoerner, L.; Mielke, C.; et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. USA 2012, 109, E934–E943. [Google Scholar] [CrossRef]
- Husain, A.; Begum, N.A.; Taniguchi, T.; Taniguchi, H.; Kobayashi, M.; Honjo, T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat. Commun. 2016, 7, 10549. [Google Scholar] [CrossRef]
- Dykhuizen, E.C.; Hargreaves, D.C.; Miller, E.L.; Cui, K.; Korshunov, A.; Kool, M.; Pfister, S.; Cho, Y.J.; Zhao, K.; Crabtree, G.R. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature 2013, 497, 624–627. [Google Scholar] [CrossRef]
- Wood, J.G.; Helfand, S.L. Chromatin structure and transposable elements in organismal aging. Front. Genet. 2013, 4, 274. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Brower-Toland, B.; Elgin, S.C. The contradictory definitions of heterochromatin: Transcription and silencing. Chromosoma 2006, 115, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Trojer, P.; Reinberg, D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol. Cell 2007, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Kouzine, F.; Levens, D. DNA topoisomerases beyond the standard role. Transcription 2013, 4, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R. The Roles of DNA Topoisomerase IIβ in Transcription. Int. J. Mol. Sci. 2018, 19, 1917. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Chan, N.L.; Hsieh, T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013, 82, 139–170. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Mouse Genome Sequencing, C.; Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Munoz-Lopez, M.; Garcia-Perez, J.L. DNA transposons: Nature and applications in genomics. Curr. Genomics 2010, 11, 115–128. [Google Scholar] [CrossRef]
- Li, W.; Jin, Y.; Prazak, L.; Hammell, M.; Dubnau, J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE 2012, 7, e44099. [Google Scholar] [CrossRef] [PubMed]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Solyom, S.; Ewing, A.D.; Rahrmann, E.P.; Doucet, T.; Nelson, H.H.; Burns, M.B.; Harris, R.S.; Sigmon, D.F.; Casella, A.; Erlanger, B.; et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012, 22, 2328–2338. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef]
- Hall, I.M.; Shankaranarayana, G.D.; Noma, K.; Ayoub, N.; Cohen, A.; Grewal, S.I. Establishment and maintenance of a heterochromatin domain. Science 2002, 297, 2232–2237. [Google Scholar] [CrossRef]
- Allshire, R. Molecular biology. RNAi and heterochromatin—A hushed-up affair. Science 2002, 297, 1818–1819. [Google Scholar] [CrossRef][Green Version]
- Villeponteau, B. The heterochromatin loss model of aging. Exp Gerontol 1997, 32, 383–394. [Google Scholar] [CrossRef]
- Larson, K.; Yan, S.J.; Tsurumi, A.; Liu, J.; Zhou, J.; Gaur, K.; Guo, D.; Eickbush, T.H.; Li, W.X. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012, 8, e1002473. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Suzuki, K.; Qu, J.; Wang, P.; Zhou, J.; Liu, X.; Ren, R.; Xu, X.; Ocampo, A.; et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 2015, 348, 1160–1163. [Google Scholar] [CrossRef]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef]
- Wang, J.; Jia, S.T.; Jia, S. New Insights into the Regulation of Heterochromatin. Trends Genet 2016, 32, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Kudlow, B.A.; Kennedy, B.K.; Monnat, R.J., Jr. Werner and Hutchinson-Gilford progeria syndromes: Mechanistic basis of human progeroid diseases. Nat. Rev. Mol. Cell Biol. 2007, 8, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Koldobskiy, M.A.; Gondor, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef]
- Dialynas, G.K.; Vitalini, M.W.; Wallrath, L.L. Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutat. Res. 2008, 647, 13–20. [Google Scholar] [CrossRef]
- Rondinelli, B.; Rosano, D.; Antonini, E.; Frenquelli, M.; Montanini, L.; Huang, D.; Segalla, S.; Yoshihara, K.; Amin, S.B.; Lazarevic, D.; et al. Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. J. Clin. Investig. 2016, 126, 4387. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Tenreiro, S.; Eckermann, K.; Outeiro, T.F. Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 2014, 7, 42. [Google Scholar] [CrossRef]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef]
- Mansuroglu, Z.; Benhelli-Mokrani, H.; Marcato, V.; Sultan, A.; Violet, M.; Chauderlier, A.; Delattre, L.; Loyens, A.; Talahari, S.; Begard, S.; et al. Loss of Tau protein affects the structure, transcription and repair of neuronal pericentromeric heterochromatin. Sci. Rep. 2016, 6, 33047. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, P.L.; Liu, Z.; Shulman, J.M. Tau Activates Transposable Elements in Alzheimer’s Disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef] [PubMed]
- Cheutin, T.; McNairn, A.J.; Jenuwein, T.; Gilbert, D.M.; Singh, P.B.; Misteli, T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 2003, 299, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Simboeck, E.; Dejardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Zuma, A.A.; Cavalcanti, D.P.; Maia, M.C.; de Souza, W.; Motta, M.C. Effect of topoisomerase inhibitors and DNA-binding drugs on the cell proliferation and ultrastructure of Trypanosoma cruzi. Int. J. Antimicrob. Agents 2011, 37, 449–456. [Google Scholar] [CrossRef]
- Baranello, L.; Bertozzi, D.; Fogli, M.V.; Pommier, Y.; Capranico, G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus. Nucleic Acids Res. 2010, 38, 159–171. [Google Scholar] [CrossRef]
- Dinh, T.T.; Gao, L.; Liu, X.; Li, D.; Li, S.; Zhao, Y.; O’Leary, M.; Le, B.; Schmitz, R.J.; Manavella, P.A.; et al. Correction: DNA Topoisomerase 1alpha Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in Arabidopsis. PLoS Genet. 2015, 11, e1005452. [Google Scholar] [CrossRef]
- Manzo, S.G.; Hartono, S.R.; Sanz, L.A.; Marinello, J.; De Biasi, S.; Cossarizza, A.; Capranico, G.; Chedin, F. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018, 19, 100. [Google Scholar] [CrossRef]
- Nakama, M.; Kawakami, K.; Kajitani, T.; Urano, T.; Murakami, Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 2012, 17, 218–233. [Google Scholar] [CrossRef]
- Kas, E.; Laemmli, U.K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992, 11, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Swedlow, J.R.; Sedat, J.W.; Agard, D.A. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell 1993, 73, 97–108. [Google Scholar] [CrossRef]
- Mengoli, V.; Bucciarelli, E.; Lattao, R.; Piergentili, R.; Gatti, M.; Bonaccorsi, S. The analysis of mutant alleles of different strength reveals multiple functions of topoisomerase 2 in regulation of Drosophila chromosome structure. PLoS Genet. 2014, 10, e1004739. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.E.; Hawley, R.S. Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet. 2014, 10, e1004650. [Google Scholar] [CrossRef] [PubMed]
- Blattes, R.; Monod, C.; Susbielle, G.; Cuvier, O.; Wu, J.H.; Hsieh, T.S.; Laemmli, U.K.; Kas, E. Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J. 2006, 25, 2397–2408. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Luke, M.; Laemmli, U.K. Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell 1991, 64, 137–148. [Google Scholar] [CrossRef]
- Jiao, W.; Lin, H.M.; Timmons, J.; Nagaich, A.K.; Ng, S.W.; Misteli, T.; Rane, S.G. E2F-dependent repression of topoisomerase II regulates heterochromatin formation and apoptosis in cells with melanoma-prone mutation. Cancer Res. 2005, 65, 4067–4077. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Jiang, L.; Lin, X.; Zhang, C.; Ou, X.; Osabe, K.; Liu, B. Changes in DNA methylation and transgenerational mobilization of a transposable element (mPing) by the topoisomerase II inhibitor, etoposide, in rice. BMC Plant Biol. 2012, 12, 48. [Google Scholar] [CrossRef]
- Hagan, C.R.; Sheffield, R.F.; Rudin, C.M. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 2003, 35, 219–220. [Google Scholar] [CrossRef]
- Miller, E.L.; Hargreaves, D.C.; Kadoch, C.; Chang, C.Y.; Calarco, J.P.; Hodges, C.; Buenrostro, J.D.; Cui, K.; Greenleaf, W.J.; Zhao, K.; et al. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 2017, 24, 344–352. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Williams, R.T.; Calarco, J.P.; Miller, E.L.; Weber, C.M.; Braun, S.M.; Pulice, J.L.; Chory, E.J.; Crabtree, G.R. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017, 49, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Xue, Y.; Shen, W.; Zhang, Y.; Joo, Y.; Ahmad, M.; Chinen, M.; Ding, Y.; Ku, W.L.; De, S.; et al. Topoisomerase 3β interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun. 2018, 9, 4946. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Shen, W.; Li, W.; Xue, Y.; Zou, S.; Xu, D.; Wang, W. Topoisomerase 3β is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res. 2017, 45, 2704–2713. [Google Scholar] [PubMed]
- Ahmad, M.; Xue, Y.; Lee, S.K.; Martindale, J.L.; Shen, W.; Li, W.; Zou, S.; Ciaramella, M.; Debat, H.; Nadal, M.; et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016, 44, 6335–6349. [Google Scholar] [CrossRef]
- Fagegaltier, D.; Bouge, A.L.; Berry, B.; Poisot, E.; Sismeiro, O.; Coppee, J.Y.; Theodore, L.; Voinnet, O.; Antoniewski, C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. USA 2009, 106, 21258–21263. [Google Scholar] [CrossRef]
- Csink, A.K.; Linsk, R.; Birchler, J.A. The Lighten up (Lip) gene of Drosophila melanogaster, a modifier of retroelement expression, position effect variegation and white locus insertion alleles. Genetics 1994, 138, 153–163. [Google Scholar]
- Gracheva, E.; Dus, M.; Elgin, S.C. Drosophila RISC component VIG and its homolog Vig2 impact heterochromatin formation. PLoS ONE 2009, 4, e6182. [Google Scholar] [CrossRef][Green Version]
- Pal-Bhadra, M.; Leibovitch, B.A.; Gandhi, S.G.; Chikka, M.R.; Bhadra, U.; Birchler, J.A.; Elgin, S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 2004, 303, 669–672. [Google Scholar] [CrossRef]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Siaw, G.E.; Willcox, S.; Griffith, J.D.; Hsieh, T.S. Synthesis and dissolution of hemicatenanes by type IA DNA topoisomerases. Proc. Natl. Acad. Sci. USA 2013, 110, E3587–E3594. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Hickson, I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 2003, 426, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Goulet, I.; Boisvenue, S.; Mokas, S.; Mazroui, R.; Cote, J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 2008, 17, 3055–3074. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Plottner, O.; Kroiss, M.; Hartmann, E.; Laggerbauer, B.; Meister, G.; Keidel, E.; Fischer, U. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 2008, 17, 3236–3246. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, W.; Zhang, L.R.; Zhang, C.Y. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol. Biosyst. 2014, 10, 1757–1764. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, H.H.; Kuwano, Y.; Abdelmohsen, K.; Srikantan, S.; Subaran, S.S.; Gleichmann, M.; Mughal, M.R.; Martindale, J.L.; Yang, X.; et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010, 17, 732–739. [Google Scholar] [CrossRef]
- Kenny, P.J.; Zhou, H.; Kim, M.; Skariah, G.; Khetani, R.S.; Drnevich, J.; Arcila, M.L.; Kosik, K.S.; Ceman, S. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014, 9, 1729–1741. [Google Scholar] [CrossRef]
- Skariah, G.; Seimetz, J.; Norsworthy, M.; Lannom, M.C.; Kenny, P.J.; Elrakhawy, M.; Forsthoefel, C.; Drnevich, J.; Kalsotra, A.; Ceman, S. Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biol. 2017, 15, 54. [Google Scholar] [CrossRef]
- Orr, W.C. Tightening the connection between transposable element mobilization and aging. Proc. Natl. Acad. Sci. USA 2016, 113, 11069–11070. [Google Scholar] [CrossRef]
- Xu, B.; Ionita-Laza, I.; Roos, J.L.; Boone, B.; Woodrick, S.; Sun, Y.; Levy, S.; Gogos, J.A.; Karayiorgou, M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012, 44, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; Ronemus, M.; Levy, D.; Wang, Z.; Hakker, I.; Rosenbaum, J.; Yamrom, B.; Lee, Y.H.; Narzisi, G.; Leotta, A.; et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012, 74, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, C.S.; Genovese, A.; Butler, M.G. Deletion of TOP3B Is Associated with Cognitive Impairment and Facial Dysmorphism. Cytogenet. Genome Res. 2016, 150, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Daghsni, M.; Lahbib, S.; Fradj, M.; Sayeb, M.; Kelmemi, W.; Kraoua, L.; Kchaou, M.; Maazoul, F.; Echebbi, S.; Ben Ali, N.; et al. TOP3B: A Novel Candidate Gene in Juvenile Myoclonic Epilepsy? Cytogenet. Genome Res. 2018, 154, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Dambacher, S.; Schotta, G. Heterochromatin dysregulation in human diseases. J. Appl. Physiol. 2010, 109, 232–242. [Google Scholar] [CrossRef]
- Muotri, A.R.; Marchetto, M.C.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature 2010, 468, 443–446. [Google Scholar] [CrossRef]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef]
- Li, W.; Prazak, L.; Chatterjee, N.; Gruninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef]
- Neale, B.M.; Kou, Y.; Liu, L.; Ma’ayan, A.; Samocha, K.E.; Sabo, A.; Lin, C.F.; Stevens, C.; Wang, L.S.; Makarov, V.; et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012, 485, 242–245. [Google Scholar] [CrossRef]
- O’Roak, B.J.; Vives, L.; Girirajan, S.; Karakoc, E.; Krumm, N.; Coe, B.P.; Levy, R.; Ko, A.; Lee, C.; Smith, J.D.; et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012, 485, 246–250. [Google Scholar] [CrossRef]
- Koga, M.; Ishiguro, H.; Yazaki, S.; Horiuchi, Y.; Arai, M.; Niizato, K.; Iritani, S.; Itokawa, M.; Inada, T.; Iwata, N.; et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet. 2009, 18, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Loe-Mie, Y.; Lepagnol-Bestel, A.M.; Maussion, G.; Doron-Faigenboim, A.; Imbeaud, S.; Delacroix, H.; Aggerbeck, L.; Pupko, T.; Gorwood, P.; Simonneau, M.; et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet. 2010, 19, 2841–2857. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.K.; Wang, W. Roles of Topoisomerases in Heterochromatin, Aging, and Diseases. Genes 2019, 10, 884. https://doi.org/10.3390/genes10110884
Lee SK, Wang W. Roles of Topoisomerases in Heterochromatin, Aging, and Diseases. Genes. 2019; 10(11):884. https://doi.org/10.3390/genes10110884
Chicago/Turabian StyleLee, Seung Kyu, and Weidong Wang. 2019. "Roles of Topoisomerases in Heterochromatin, Aging, and Diseases" Genes 10, no. 11: 884. https://doi.org/10.3390/genes10110884
APA StyleLee, S. K., & Wang, W. (2019). Roles of Topoisomerases in Heterochromatin, Aging, and Diseases. Genes, 10(11), 884. https://doi.org/10.3390/genes10110884



