RNA Sequencing Reveals That Both Abiotic and Biotic Stress-Responsive Genes are Induced during Expression of Steroidal Glycoalkaloid in Potato Tuber Subjected to Light Exposure
Abstract
:1. Introduction
2. Methods
2.1. Biological Materials and Treatment
2.2. RNA Extraction, RNA-Seq Library Construction and Sequencing
2.3. Analysis of Sequencing Data
2.4. Cis-Element Prediction and Weighted Gene Co-Expression Network Analysis (WGCNA)
2.5. Quantitative Real-Time PCR
3. Results
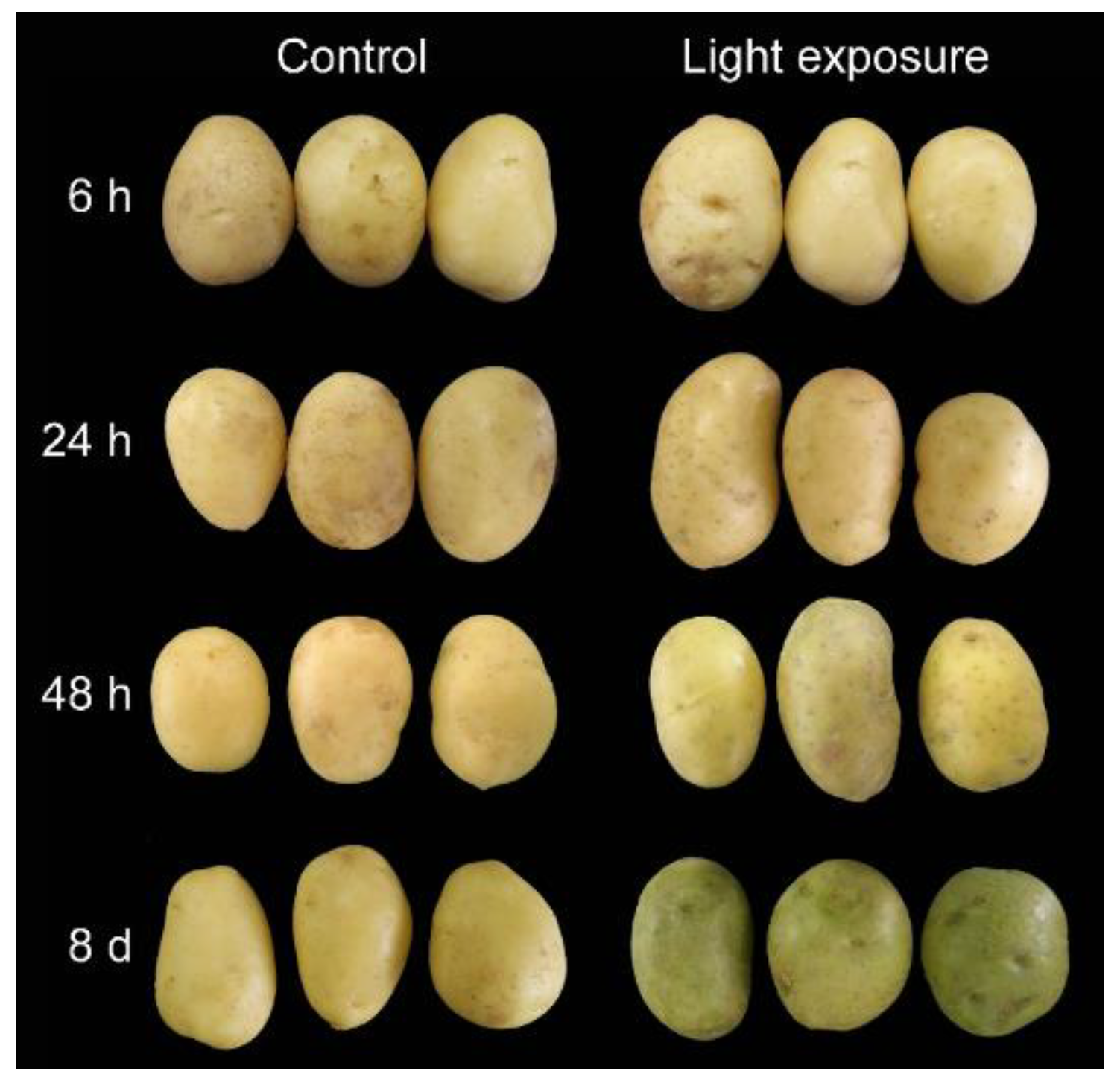
3.1. Potato Tubers with a Difference in Phenotypes
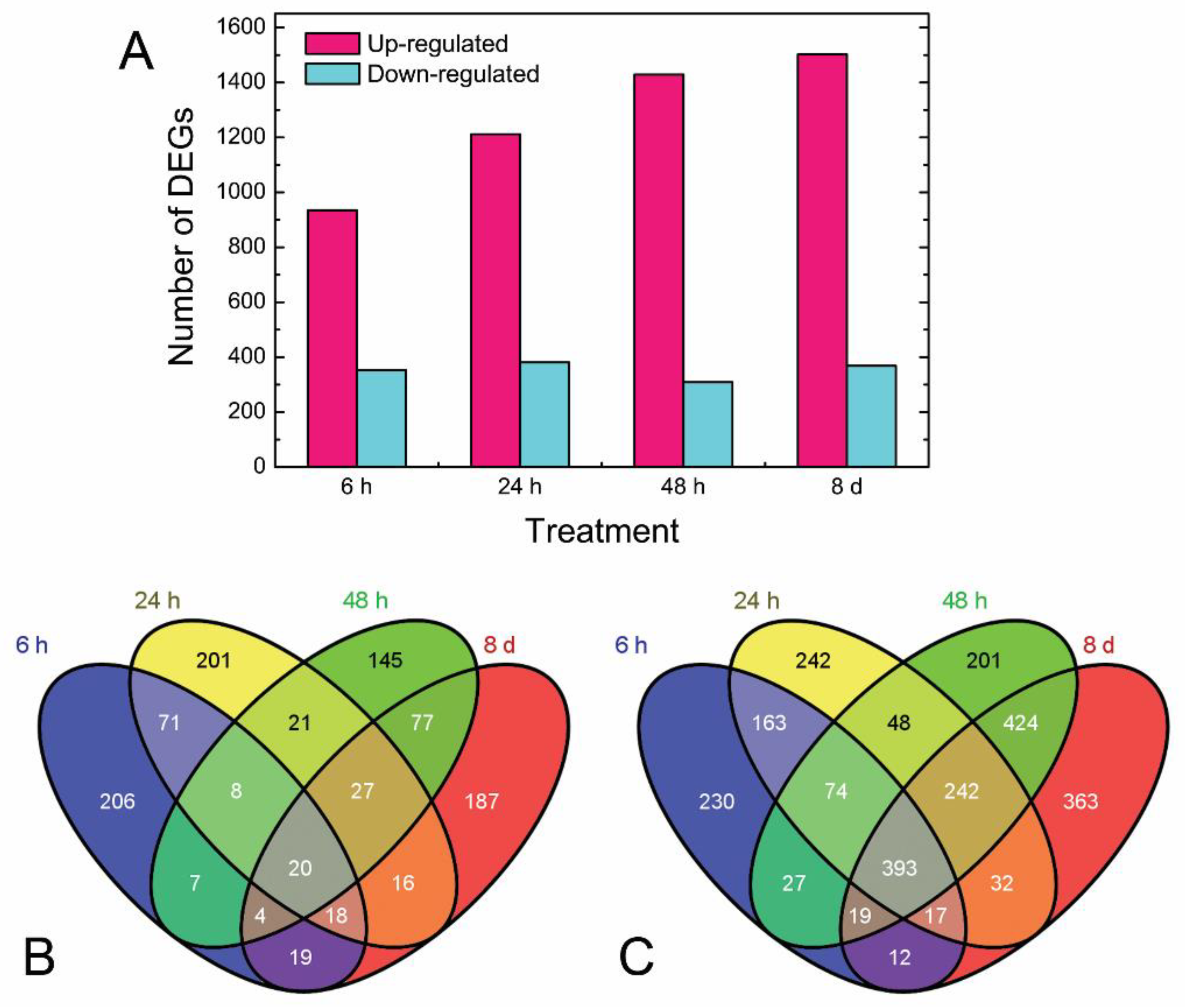
3.2. Quality of Data and Differentially Expressed Genes
3.3. Expression Patterns of the Steroidal Glycoalkaloid (SGA)-Related Genes
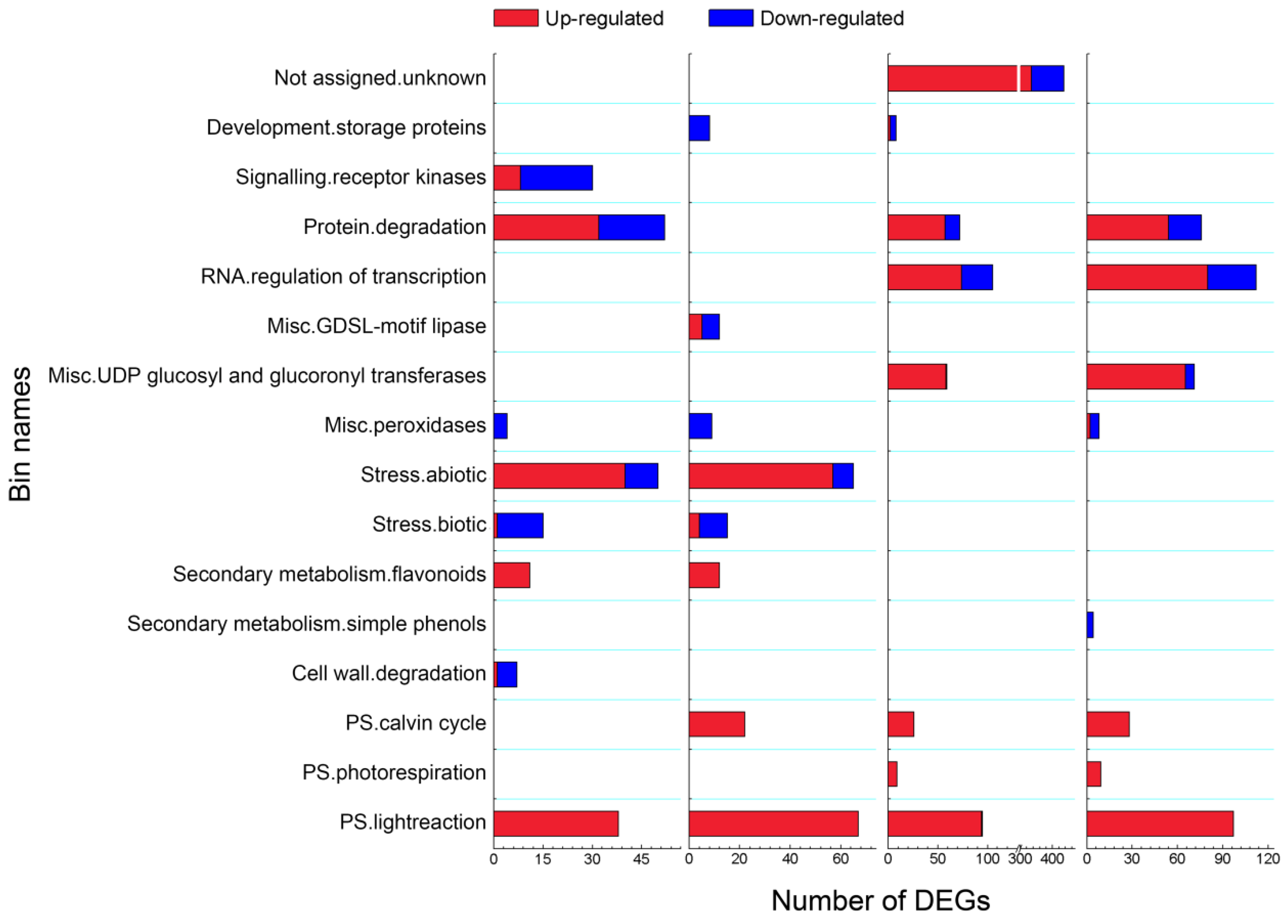
3.4. Overview of Mapman Analysis
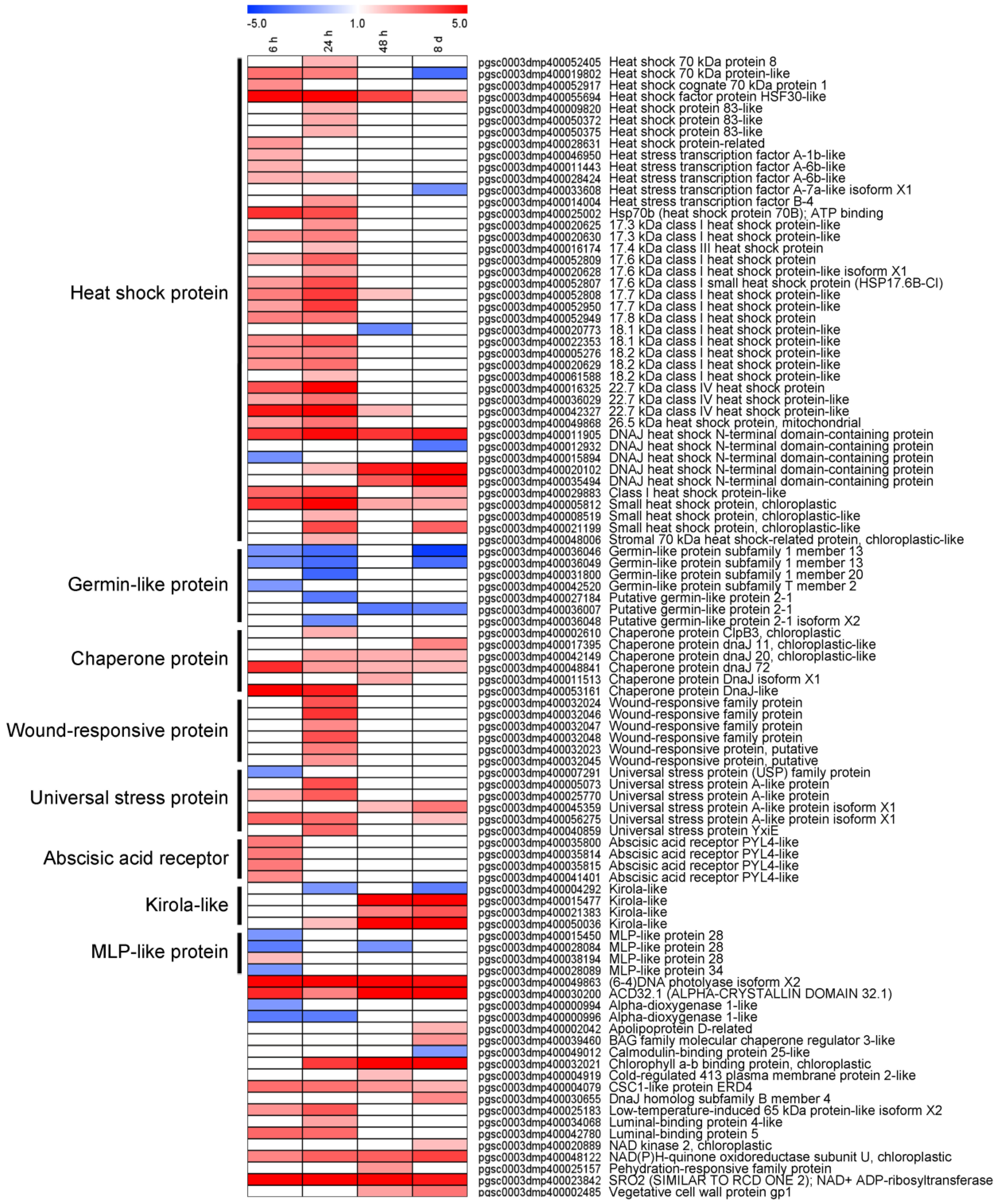
3.5. DEGs Involved in Abiotic Stress Were Differentially Expressed in Response to Light Exposure
3.6. DEGs Involved in Biotic Stress Were Differentially Expressed in Response to Light Exposure
3.7. Cis-Elements in the Promoter Region of SGA-Biosynthetic and Disease-Resistant Genes
3.8. Verification of DEGs Using qRT-PCR
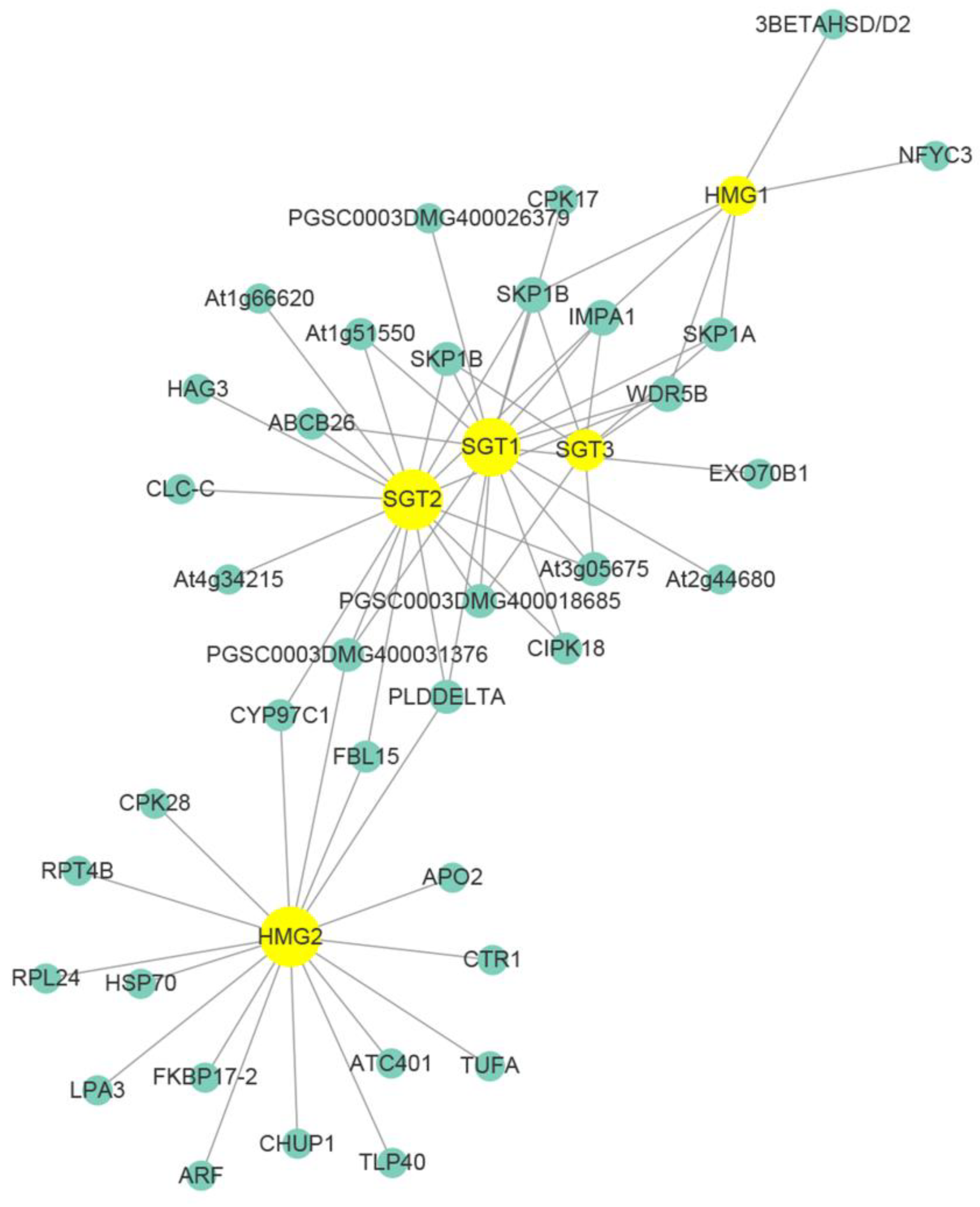
3.9. Co-Expressional Networks of Genes between SGA and Disease Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schieber, A.; Aranda Saldaña Marleny, D. Potato peels: A source of nutritionally and pharmacologically interesting compounds-A review. Food 2009, 3, 23–29. [Google Scholar]
- Friedman, M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J. Agric. Food Chem. 2006, 54, 8655–8681. [Google Scholar] [CrossRef] [PubMed]
- Nahar, N.; Westerberg, E.; Arif, U.; Huchelmann, A.; Olarte Guasca, A.; Beste, L.; Dalman, K.; Dutta, P.C.; Jonsson, L.; Sitbon, F. Transcript profiling of two potato cultivars during glycoalkaloid-inducing treatments shows differential expression of genes in sterol and glycoalkaloid metabolism. Sci. Rep. 2017, 7, 43268. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.J.; Sharma, R.P.; Salunkhe, D.K. Naturally occurring toxic alkaloids in foods. Crit. Rev. Toxicol. 1981, 9, 21–104. [Google Scholar] [CrossRef] [PubMed]
- Sanford, L.L.; Deahl, K.L.; Sinden, S.L.; Ladd, T.L. Glycoalkaloid contents in tubers from Solanum tuberosum populations selected for potato leafhopper resistance. Am. J. Potato Res. 1992, 69, 693–703. [Google Scholar] [CrossRef]
- Andreu, A.; Oliva, C.; Distel, S.; Daleo, G. Production of phytoalexins, glycoalkaloidsand phenolics in leaves and tubers of potato cultivars with different degrees of field resistance after infection with Phytophthora infestans. Potato Res. 2001, 44, 1–9. [Google Scholar] [CrossRef]
- Sarquís, J.I.; Coria, N.A.; Aguilar, I.; Rivera, A. Glycoalkaloid content in Solanum species and hybrids from a breeding program for resistance to late blight (Phytophthora infestans). Am. J. Potato Res. 2000, 77, 295–302. [Google Scholar] [CrossRef]
- Watson, A.A.; Davies, D.R.; Asano, N.; Winchester, B.; Kato, A.; Molyneux, R.J.; Stegelmeier, B.L.; Nash, R.J. Calystegine alkaloids in the potato and other food plants. In Natural and Selected Synthetic Toxins; Tu, A.T., Gaffield, W., Eds.; American Chemical Society: Washington, DC, USA, 2000; pp. 129–139. [Google Scholar]
- Dale, M.F.; Griffiths, D.W.; Bain, H.; Todd, D. Glycoalkaloid increase in Solarium tuberosum on exposure to light. Ann. Appl. Biol. 1993, 123, 411–418. [Google Scholar] [CrossRef]
- Ginzberg, I.; Tokuhisa, J.G.; Veilleux, R.E. Potato steroidal glycoalkaloids: Biosynthesis and genetic manipulation. Potato Res. 2009, 52, 1–15. [Google Scholar] [CrossRef]
- Taylor, M.A.; Mcdougall, G.J.; Stewart, D. Potato flavor and texture. In Potato Biology and Biotechnology; Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., Taylor, M.A., MacKerron, D.K.L., Ross, H.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 525–540. [Google Scholar]
- Dale, M.F.; Griffiths, D.W.; Bain, H. Effect of bruising on the total glycoalkaloid and chlorogenic acid content of potato (Solanum tuberosum) tubers of five cultivars. J. Sci. Food Agric. 1998, 77, 499–505. [Google Scholar] [CrossRef]
- Friedman, M.; Mcdonald, G.M. Postharvest changes in glycoalkaloid content of potatoes. Adv. Exp. Med. Biol. 1999, 459, 121–143. [Google Scholar] [PubMed]
- Haase, N.U. Glycoalkaloid concentration in potato tubers related to storage and consumer offering. Potato Res. 2010, 53, 297–307. [Google Scholar] [CrossRef]
- Dao, L.; Friedman, M. Chlorophyll, chlorogenic acid, glycoalkaloid, and protease inhibitor content of fresh and green potatoes. J. Agric. Food Chem. 1994, 42, 633–639. [Google Scholar] [CrossRef]
- Percival, G.C.; Karim, M.S. Influence of light-enhanced glycoalkaloids on resistance of potato tubers to fusarium sulphureum and fusarium solani var. coeruleum. Plant Pathol. 2010, 47, 665–670. [Google Scholar] [CrossRef]
- Ohyama, K.; Okawa, A.; Moriuchi, Y.; Fujimoto, Y. Biosynthesis of steroidal alkaloids in Solanaceae plants: Involvement of an aldehyde during C-26 amination. Phytochemistry 2013, 89, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Petersson, E.V.; Nurun, N.; Paul, D.; Anders, B.; Rikard, T.; Dutta, P.C.; Jonsson, L.; Sitbon, F. Conversion of exogenous cholesterol into glycoalkaloids in potato shoots, using two methods for sterol solubilisation. PLoS ONE 2013, 8, e82955. [Google Scholar] [CrossRef] [PubMed]
- Sitbon, F.; Jonsson, L. Sterol composition and growth of transgenic tobacco plants expressing type-1 and type-2 sterol methyltransferases. Planta 2001, 212, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Arnqvist, L.; Dutta, P.C.; Jonsson, L.; Sitbon, F. Reduction of cholesterol and glycoalkaloid levels in transgenic potato plants by overexpression of a type 1 sterol methyltransferase cDNA. Plant. Physiol. 2003, 131, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
- Carpintero, M.; Constanza, N. Genetic Studies of Candidate Genes in the Glycoalkaloid Biosynthetic Pathway of Potato. Ph.D. Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, December 2012. [Google Scholar]
- Ginzberg, I.; Fogelman, E.; Demirel, U.; Mweetwa, A.M.; Tokuhisa, J.; Veilleux, R.E. Induction of potato steroidal glycoalkaloid biosynthetic pathway by overexpression of cDNA encoding primary metabolism HMG-CoA reductase and squalene synthase. Planta 2012, 235, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lou, Y.; Sun, H.; Li, L.; Wang, L.; Dong, L.; Gao, Z. Transcriptome and comparative gene expression analysis of Phyllostachys edulis in response to high light. BMC Plant. Biol. 2016, 16, 34. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Kim, H.Y.; Kim, H.S.; Jung, S.H. Transcriptome analysis of Panax ginseng response to high light stress. J. Ginseng Res. 2019. [Google Scholar] [CrossRef]
- Deng, Y.; Yao, J.; Wang, X.; Guo, H.; Duan, D. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS ONE 2012, 7, e39704. [Google Scholar] [CrossRef] [PubMed]
- Zuo, C.; Zhang, W.; Chen, Z.; Chen, B.; Huang, Y. RNA sequencing reveals that endoplasmic reticulum stress and disruption of membrane integrity underlie dimethyl trisulfide toxicity against Fusarium oxysporum f. sp. cubense tropical Race 4. Front. Microbiol. 2017, 8, 1365. [Google Scholar] [CrossRef] [PubMed]
- Hardigan, M.A.; Crisovan, E.; Hamilton, J.P.; Kim, J.; Laimbeer, P.; Leisner, C.P.; Manrique-Carpintero, N.C.; Newton, L.; Pham, G.M.; Vaillancourt, B.; et al. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 2016, 28, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2010, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Moreau, Y.; De Moor, B.; Rouzé, P.; Rombauts, S. PlantCARE: A database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Manrique-Carpintero, N.C.; Tokuhisa, J.G.; Idit, G.; Holliday, J.A.; Veilleux, R.E. Sequence diversity in coding regions of candidate genes in the glycoalkaloid biosynthetic pathway of wild potato species. G3 Genes Genomes Genet. 2013, 3, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yan, Z.; Xu, Z.; Wang, Y.; Xie, Z. Transcriptome analysis and physiological responses of the potato plantlets in vitro under red, blue, and white light conditions. 3 Biotech 2018, 8, 394. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, D.W.; Bain, H.; Dale, M.B. Effect of storage temperature on potato (Solanum tuberosum L.) tuber glycoalkaloid content and the subsequent accumulation of glycoalkaloids and chlorophyll in response to light exposure. J. Agric. Food Chem. 1998, 46, 5262–5268. [Google Scholar] [CrossRef]
- Gull, D.D.; Isenberg, F.M. Chlorophyll and solanine content and distribution in four varieties of potato tubers. Proc. Am. Soc. Hortic. Sci. 1960, 75, 545–556. [Google Scholar]
- Edwards, E.J.; Saint, R.E.; Cobb, A.H. Is there a link between greening and light-enhanced glycoalkaloid accumulation in potato (Solanum tuberosum L.) tubers? J. Sci. Food Agric. 1998, 76, 327–333. [Google Scholar] [CrossRef]
- Andrivon, D.; Corbiere, R.; Lucas, J.M.; Pasco, C.; Gravoueille, J.M.; Pelle, R.; Dantec, J.P.; Ellissèche, D. Resistance to late blight and soft rot in six potato progenies and glycoalkaloid contents in the tubers. Am. J. Potato Res. 2003, 80, 125–134. [Google Scholar] [CrossRef]
- Birch, A.E.; Geoghegan, I.E.; Griffiths, D.W.; Mcnicol, J.W. The effect of genetic transformations for pest resistance on foliar solanidine-based glycoalkaloids of potato (Solatium tuberosuni). Ann. Appl. Biol. 2015, 140, 143–149. [Google Scholar] [CrossRef]
- Calf, O.W.; Huber, H.; Peters, J.L.; Weinhold, A.; van Dam, N.M. Glycoalkaloid composition explains variation in slug resistance in solanum dulcamara. Oecologia 2018, 187, 495–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, P.; Müller Marion, C.; Ekengren, S.; Mckee, L.S.; Bulone, V. The impact of steroidal glycoalkaloids on the physiology infestans, the causative agent of potato late blight. Mol. Plant Microbe Interact. 2017, 30, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Mweetwa, A.M.; Hunter, D.; Poe, R.; Harich, K.C.; Ginzberg, I.; Veilleux, R.E.; Tokuhisa, J.G. Steroidal glycoalkaloids in solanum chacoense. Phytochemistry 2012, 75, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Maldonado, A.F.; Schieber, A.; Gänzle, M.G. Antifungal activity of secondary plant metabolites from potatoes (Solanum tuberosum): Glycoalkaloids and phenolic acids show synergistic effects. J. Appl. Microbiol. 2016, 120, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Petersson, E.V.; Arif, U.; Schulzova, V.; Krtková, V.; Hajšlová, J.; Meijer, J.; Andersson, H.C.; Jonsson, L.; Sitbon, F. Glycoalkaloid and calystegine levels in table potato cultivars subjected to wounding, light, and heat treatments. J. Agric. Food Chem. 2013, 61, 5893–5902. [Google Scholar] [CrossRef] [PubMed]
- Lewinsohn, E.; Gijzen, M.; Barton, K.; Croteau, R. Oleoresinosis in grand fir (Abies grandis) saplings and mature trees: Modulation of this wound response by light and water stresses. Plant. Physiol. 1993, 101, 1021–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Rokka, V.M.; Laurila, J.; Tauriainen, A.; Laakso, I.; Larkka, J.; Metzler, M.; Pietilä, L. Glycoalkaloid aglycone accumulations associated with infection by Clavibacter michiganensis ssp. sepedonicus in potato species Solanum acaule and Solanum tuberosum and their interspecific somatic hybrids. Plant Cell Rep. 2005, 23, 683–691. [Google Scholar] [PubMed]
- Burton, H.R.; Kuć, J. The effect of sterols on phytoalexin, steroid glycoalkaloid, and sterol accumulation in potato tuber discs inoculated with Phytophthora infestans, or treated with arachidonic acid. Physiol. Mol. Plant Pathol. 1995, 47, 13–27. [Google Scholar]
- Shinde, B.A.; Dholakia, B.B.; Hussain, K.; Panda, S.; Meir, S.; Rogachev, I.; Aharoni, A.; Giri, A.P.; Kamble, A.C. Dynamic metabolic reprogramming of steroidal glycol-alkaloid and phenylpropanoid biosynthesis may impart early blight resistance in wild tomato (Solanum arcanum Peralta). Plant Mol. Biol. 2017, 95, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sleckman, B.P.; Gorman, J.R.; Alt, F.W. Accessibility control of antigen-receptor variable-region gene assembly: Role of cis-acting elements. Annu. Rev. Immunol. 1996, 14, 459–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derksen, H.; Rampitsch, C.; Daayf, F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013, 207, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Vidhyasekaran, P. Plant Hormone Signaling Systems in Plant Innate Immunity; Springer: Berlin, Germany, 2015. [Google Scholar]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267. [Google Scholar] [CrossRef] [PubMed]
- Childs, K.L.; Davidson, R.M.; Buell, C.R. Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS ONE 2011, 6, e22196. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Sample | Raw Reads | Clean Reads | Clean Data Ratio (%) | Mapped Genome (%) | Mapped Gene (%) | Expressed Gene |
|---|---|---|---|---|---|---|
| C1-1 | 42453718 | 40692256 | 95.85 | 75.26% | 61.32% | 21502 |
| C1-2 | 42453868 | 40784058 | 96.07 | 75.65% | 62.10% | 21760 |
| C1-3 | 42453938 | 40851000 | 96.22 | 77.15% | 63.42% | 21234 |
| C2-1 | 42452090 | 40489950 | 95.38 | 78.06% | 63.31% | 21213 |
| C2-2 | 42449108 | 40526196 | 95.47 | 77.66% | 63.54% | 21240 |
| C2-3 | 42450246 | 40465220 | 95.32 | 77.99% | 63.37% | 21296 |
| C3-1 | 42452696 | 40184268 | 94.66 | 79.00% | 62.55% | 20684 |
| C3-2 | 42454638 | 40396030 | 95.15 | 79.99% | 64.05% | 20583 |
| C3-3 | 42453942 | 40565054 | 95.55 | 78.69% | 62.36% | 20724 |
| C4-1 | 42453418 | 40439256 | 95.26 | 79.97% | 62.67% | 20343 |
| C4-2 | 42454958 | 40429838 | 95.23 | 80.43% | 64.14% | 20208 |
| C4-3 | 44085796 | 41531086 | 94.21 | 80.20% | 64.30% | 20317 |
| T1-1 | 42452422 | 40900082 | 96.34 | 78.26% | 64.01% | 21252 |
| T1-2 | 42453514 | 40644482 | 95.74 | 78.82% | 65.30% | 21497 |
| T1-3 | 42453450 | 40262838 | 94.84 | 74.38% | 61.61% | 21439 |
| T2-1 | 42452342 | 40865858 | 96.26 | 77.43% | 63.81% | 21383 |
| T2-2 | 42452858 | 40092998 | 94.44 | 74.45% | 62.40% | 21523 |
| T2-3 | 42452510 | 40548506 | 95.51 | 75.35% | 62.90% | 21296 |
| T3-1 | 42452560 | 40916514 | 96.38 | 78.03% | 65.32% | 21088 |
| T3-2 | 42454240 | 40920104 | 96.39 | 76.46% | 64.75% | 20893 |
| T3-3 | 42454122 | 40547356 | 95.51 | 75.17% | 63.14% | 20942 |
| T4-1 | 42453850 | 40831056 | 96.18 | 74.59% | 62.51% | 20767 |
| T4-2 | 42453644 | 40765702 | 96.02 | 75.71% | 63.58% | 20667 |
| T4-3 | 42453510 | 40945040 | 96.45 | 76.61% | 65.58% | 20828 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Zuo, C.; Chen, Z.; Kang, Y.; Qin, S. RNA Sequencing Reveals That Both Abiotic and Biotic Stress-Responsive Genes are Induced during Expression of Steroidal Glycoalkaloid in Potato Tuber Subjected to Light Exposure. Genes 2019, 10, 920. https://doi.org/10.3390/genes10110920
Zhang W, Zuo C, Chen Z, Kang Y, Qin S. RNA Sequencing Reveals That Both Abiotic and Biotic Stress-Responsive Genes are Induced during Expression of Steroidal Glycoalkaloid in Potato Tuber Subjected to Light Exposure. Genes. 2019; 10(11):920. https://doi.org/10.3390/genes10110920
Chicago/Turabian StyleZhang, Weina, Cunwu Zuo, Zhongjian Chen, Yichen Kang, and Shuhao Qin. 2019. "RNA Sequencing Reveals That Both Abiotic and Biotic Stress-Responsive Genes are Induced during Expression of Steroidal Glycoalkaloid in Potato Tuber Subjected to Light Exposure" Genes 10, no. 11: 920. https://doi.org/10.3390/genes10110920
APA StyleZhang, W., Zuo, C., Chen, Z., Kang, Y., & Qin, S. (2019). RNA Sequencing Reveals That Both Abiotic and Biotic Stress-Responsive Genes are Induced during Expression of Steroidal Glycoalkaloid in Potato Tuber Subjected to Light Exposure. Genes, 10(11), 920. https://doi.org/10.3390/genes10110920




