Cell-Specific DNA Methylation Signatures in Asthma
Abstract
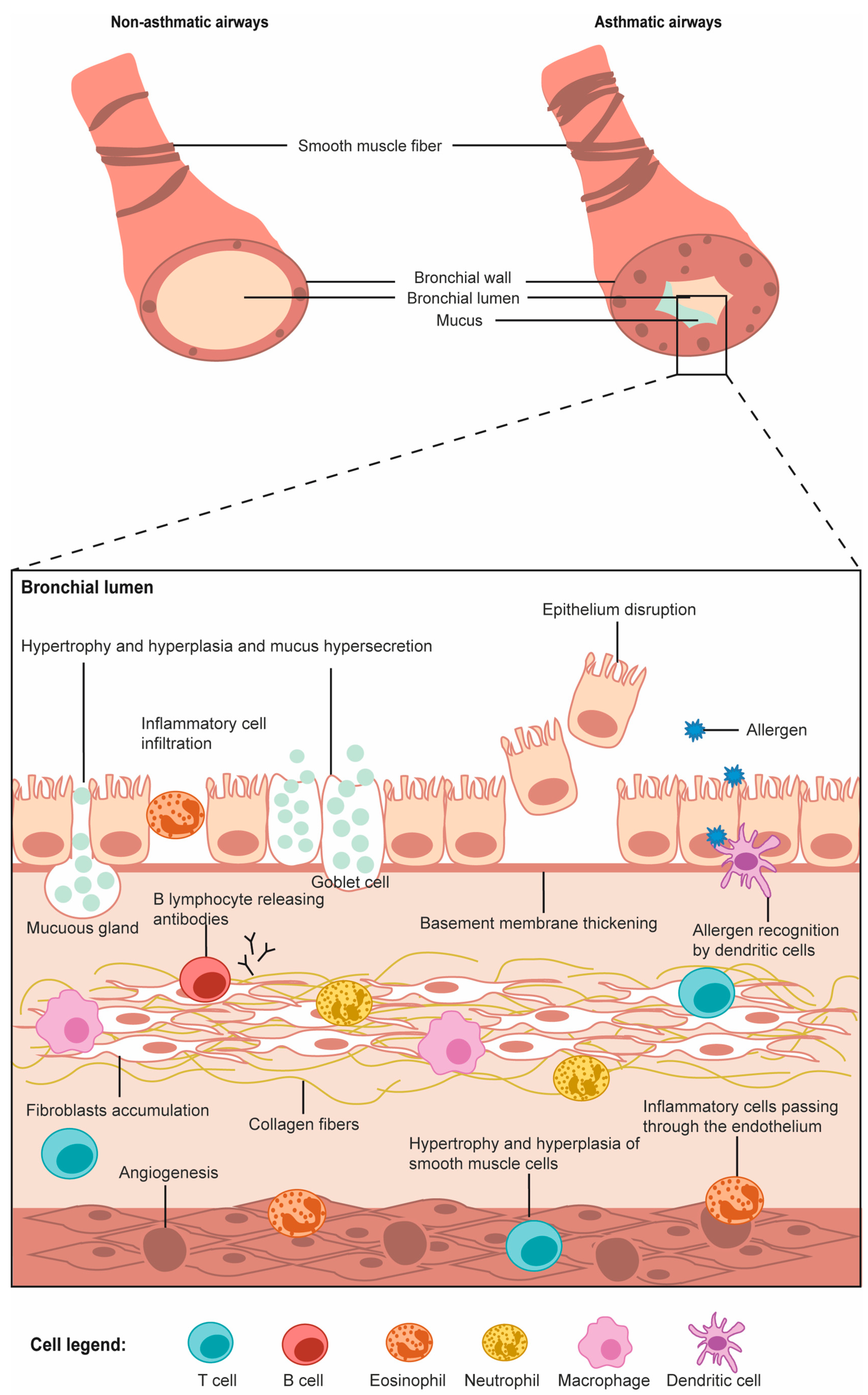
:1. Introduction
2. Cell-Specific DNA Methylation Modifications Associated with Asthma
2.1. Cells of the Immune System
2.1.1. Granulocytes: Eosinophils, Neutrophils, Basophils and Mastocytes
2.1.2. Monocytes and Macrophages
2.1.3. Dendritic Cells
2.1.4. Lymphocytes T (CD4+ and CD8+)
2.1.5. Lymphocytes B (CD19+)
2.1.6. Natural Killer Cells (NK)
2.1.7. Platelets and Megakaryocytes
2.2. Cells of the Respiratory Tract
2.2.1. Airway Epithelial Cells
2.2.2. Airway Smooth Muscle Cells
3. Differences in DNA Methylation Associated with Asthma among Various Cell Types
3.1. DNA Methylation Machinery
3.2. Comparative Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Asthma. Available online: https://www.who.int/news-room/fact-sheets/detail/asthma (accessed on 22 July 2019).
- Network, G.A. The Global Asthma Report; Global Asthma Network: Auckland, New Zealand, 2018. [Google Scholar]
- Lundbäck, B.; Backman, H.; Lötvall, J.; Rönmark, E. Is asthma prevalence still increasing? Expert Rev. Respir. Med. 2016, 10, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.R.; Stokes, K.; August, A. The role of eosinophils in allergic airway inflammation. Discov. Med. 2010, 9, 357–362. [Google Scholar] [PubMed]
- Barnes, P.J. Pathophysiology of allergic inflammation. Immunol. Rev. 2011, 242, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Sindher, S.B.; Long, A.; Acharya, S.; Sampath, V.; Nadeau, K.C. The use of biomarkers to predict aero-allergen and food immunotherapy responses. Clin. Rev. Allergy Immunol. 2018, 55, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Morishima, Y.; Ano, S.; Ishii, Y.; Ohtsuka, S.; Matsuyama, M.; Kawaguchi, M.; Hizawa, N. Th17-associated cytokines as a therapeutic target for steroid-insensitive asthma. Clin. Dev. Immunol. 2013, 2013, 609395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-T.; Wang, C.-Z. Regulatory t cells and asthma. J. Zhejiang Univ. Sci. B 2018, 19, 663–673. [Google Scholar] [CrossRef]
- Chesné, J.; Braza, F.; Mahay, G.; Brouard, S.; Aronica, M.; Magnan, A. Il-17 in severe asthma. Where do we stand? Am. J. Respir. Crit. Care Med. 2014, 190, 1094–1101. [Google Scholar] [CrossRef]
- Newcomb, D.C.; Peebles, R.S., Jr. Th17-mediated inflammation in asthma. Curr. Opin. Immunol. 2013, 25, 755–760. [Google Scholar] [CrossRef]
- Rakesh, K.K.; Ming, Y.; Cristan, H.; Paul, S.F. Interferon-γ, pulmonary macrophages and airway responsiveness in asthma. Inflamm. Allergy Drug Targets 2012, 11, 292–297. [Google Scholar]
- Moldaver, D.M.; Larché, M.; Rudulier, C.D. An update on lymphocyte subtypes in asthma and airway disease. Chest 2017, 151, 1122–1130. [Google Scholar] [CrossRef]
- Kon, O.M.; Kay, A.B. T cells and chronic asthma. Int Arch. Allergy Immunol. 1999, 118, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Possa, S.S.; Leick, E.A.; Prado, C.M.; Martins, M.A.; Tibério, I.F.L.C. Eosinophilic inflammation in allergic asthma. Front. Pharmacol. 2013, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- De Nijs, S.B.; Venekamp, L.N.; Bel, E.H. Adult-onset asthma: Is it really different? Eur. Respir. Rev. 2013, 22, 44. [Google Scholar] [CrossRef] [PubMed]
- Knight, D. Epithelium–fibroblast interactions in response to airway inflammation. Immunol. Cell Biol. 2001, 79, 160–164. [Google Scholar] [CrossRef]
- Wright, S.M.; Hockey, P.M.; Enhorning, G.; Strong, P.; Reid, K.B.M.; Holgate, S.T.; Djukanovic, R.; Postle, A.D. Altered airway surfactant phospholipid composition and reduced lung function in asthma. J. Appl. Phys. 2000, 89, 1283–1292. [Google Scholar] [CrossRef]
- Winkler, C.; Hohlfeld, J.M. Surfactant and allergic airway inflammation. Swiss Med. Wkl. 2013, 143, w13818. [Google Scholar] [CrossRef]
- Ober, C. Asthma genetics in the post-gwas era. Ann. Am. Thorac Soc. 2016, 13 (Suppl. 1), S85–S90. [Google Scholar]
- Lee, J.-U.; Kim, J.D.; Park, C.-S. Gene-environment interactions in asthma: Genetic and epigenetic effects. Yonsei Med. J. 2015, 56, 877–886. [Google Scholar] [CrossRef]
- Noutsios, G.T.; Floros, J. Childhood asthma: Causes, risks, and protective factors; a role of innate immunity. Siss Med. Wkl. 2014, 144, w14036. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics links genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Bellani, J.A. Genetics/epigenetics/allergy: The gun is loaded... But what pulls the trigger? Allergy Asthma Proc. 2019, 40, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Gen. 2003, 33, 245. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.; Jurkowski, T.P.; Kellner, S.; Schneider, D.; Jeltsch, A.; Helm, M. The rna methyltransferase dnmt2 methylates DNA in the structural context of a trna. RNA Biol. 2017, 14, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Nellen, W.; Lyko, F. Two substrates are better than one: Dual specificities for dnmt2 methyltransferases. Trends Biochem. Sci. 2006, 31, 306–308. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog dnmt2. Science 2006, 311, 395. [Google Scholar] [CrossRef]
- Schaefer, M.; Lyko, F. Solving the dnmt2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar] [CrossRef]
- Tsagaratou, A.; Lio, C.-W.J.; Yue, X.; Rao, A. Tet methylcytosine oxidases in t cell and b cell development and function. Front. Immunol. 2017, 8, 220. [Google Scholar] [CrossRef]
- Holland, N. Future of environmental research in the age of epigenomics and exposomics. Rev. Environ. Health 2017, 32, 45–54. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA hypermethylation in disease: Mechanisms and clinical relevance. Epigenetics 2019, 14, 1–23. [Google Scholar] [CrossRef]
- Mørkve Knudsen, T.; Rezwan, F.I.; Jiang, Y.; Karmaus, W.; Svanes, C.; Holloway, J.W. Transgenerational and intergenerational epigenetic inheritance in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 765–772. [Google Scholar] [CrossRef]
- Vercelli, D. Does epigenetics play a role in human asthma? Allergol. Int. 2016, 65, 123–126. [Google Scholar] [CrossRef] [PubMed]
- DeVries, A.; Vercelli, D. Epigenetic mechanisms in asthma. AnnalsATS 2016, 13, S48–S50. [Google Scholar] [PubMed]
- Davidson, E.J.; Yang, I.V. Role of epigenetics in the development of childhood asthma. Curr. Opin. Allergy Clin. Immunol. 2018, 18, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Alaskhar Alhamwe, B.; Khalaila, R.; Wolf, J.; von Bülow, V.; Harb, H.; Alhamdan, F.; Hii, C.S.; Prescott, S.L.; Ferrante, A.; Renz, H.; et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. Off. J. Can. Soc. Allergy Clin. Immunol. 2018, 14, 39. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, K.A.; Clifford, R.L.; Knox, A.J. Epigenetic changes in airway smooth muscle as a driver of airway inflammation and remodeling in asthma. Chest 2019, 155, 816–824. [Google Scholar] [CrossRef]
- Tost, J. A translational perspective on epigenetics in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 715–726. [Google Scholar] [CrossRef] [Green Version]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef]
- Farzan, N.; Vijverberg, S.J.; Kabesch, M.; Sterk, P.J.; Zee, A.H.M.v.d. The use of pharmacogenomics, epigenomics, and transcriptomics to improve childhood asthma management: Where do we stand? Pediatr. Pulmonol. 2018, 53, 836–845. [Google Scholar] [CrossRef]
- Forno, E.; Celedón, J.C. Epigenomics and transcriptomics in the prediction and diagnosis of childhood asthma: Are we there yet? Front. Pediatr. 2019, 7, 115. [Google Scholar] [CrossRef]
- Saco, T.V.; Breitzig, M.T.; Lockey, R.F.; Kolliputi, N. Epigenetics of mucus hypersecretion in chronic respiratory diseases. Am. J. Respir Cell Mol. Biol. 2018, 58, 299–309. [Google Scholar] [CrossRef]
- Zakarya, R.; Adcock, I.; Oliver, B.G. Epigenetic impacts of maternal tobacco and e-vapour exposure on the offspring lung. Clin. Epigenet. 2019, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Riedhammer, C.; Halbritter, D.; Weissert, R. Peripheral blood mononuclear cells: Isolation, freezing, thawing, and culture. In Multiple Sclerosis: Methods and Protocols; Weissert, R., Ed.; Springer: New York, NY, USA, 2016; pp. 53–61. [Google Scholar]
- Titus, A.J.; Gallimore, R.M.; Salas, L.A.; Christensen, B.C. Cell-type deconvolution from DNA methylation: A review of recent applications. Hum. Mol. Gen. 2017, 26, R216–R224. [Google Scholar] [CrossRef] [Green Version]
- The UniProt Consortium. Uniprot: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [PubMed]
- Fotis, P.; Giorgos, H.; Stelios, L. Phenotyping and endotyping asthma based on biomarkers. Curr. Top. Med. Chem. 2016, 16, 1582–1586. [Google Scholar]
- Fatemi, F.; Sadroddiny, E.; Gheibi, A.; Mohammadi Farsani, T.; Kardar, G.A. Biomolecular markers in assessment and treatment of asthma. Respirology 2014, 19, 514–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Froidure, A.; Mouthuy, J.; Durham, S.R.; Chanez, P.; Sibille, Y.; Pilette, C. Asthma phenotypes and ige responses. Eur. Respir. J. 2016, 47, 304. [Google Scholar] [CrossRef] [Green Version]
- Zissler, U.M.; Esser-von Bieren, J.; Jakwerth, C.A.; Chaker, A.M.; Schmidt-Weber, C.B. Current and future biomarkers in allergic asthma. Allergy 2016, 71, 475–494. [Google Scholar] [CrossRef]
- Edris, A.; den Dekker, H.T.; Melén, E.; Lahousse, L. Epigenome-wide association studies in asthma: A systematic review. Clin. Exp. Allergy 2019, 49, 953–968. [Google Scholar] [CrossRef]
- Xu, C.-J.; Söderhäll, C.; Bustamante, M.; Baïz, N.; Gruzieva, O.; Gehring, U.; Mason, D.; Chatzi, L.; Basterrechea, M.; Llop, S.; et al. DNA methylation in childhood asthma: An epigenome-wide meta-analysis. Lancet Respir. Med. 2018, 6, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Reese, S.E.; Xu, C.-J.; den Dekker, H.T.; Lee, M.K.; Sikdar, S.; Ruiz-Arenas, C.; Merid, S.K.; Rezwan, F.I.; Page, C.M.; Ullemar, V.; et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J. Allergy Clin. Immunol. 2019, 143, 2062–2074. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Willis-Owen, S.A.G.; Laprise, C.; Wong, K.C.C.; Davies, G.A.; Hudson, T.J.; Binia, A.; Hopkin, J.M.; Yang, I.V.; Grundberg, E.; et al. An epigenome-wide association study of total serum immunoglobulin e concentration. Nature 2015, 520, 670–674. [Google Scholar] [CrossRef] [Green Version]
- Arathimos, R.; Suderman, M.; Sharp, G.C.; Burrows, K.; Granell, R.; Tilling, K.; Gaunt, T.R.; Henderson, J.; Ring, S.; Richmond, R.C.; et al. Epigenome-wide association study of asthma and wheeze in childhood and adolescence. Clin. Epigenet. 2017, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Wang, T.; Pino-Yanes, M.; Forno, E.; Liang, L.; Yan, Q.; Hu, D.; Weeks, D.E.; Baccarelli, A.; Acosta-Perez, E.; et al. An epigenome-wide association study of total serum ige in hispanic children. J. Allergy Clin. Immunol. 2017, 140, 571–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, I.V.; Richards, A.; Davidson, E.J.; Stevens, A.D.; Kolakowski, C.A.; Martin, R.J.; Schwartz, D.A. The nasal methylome: A key to understanding allergic asthma. Am. J. Respir. Crit. Care Med. 2017, 195, 829–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, A.; Urbanek, C.; Eng, C.; Schageman, J.; Jacobson, S.; O’Connor, B.P.; Galanter, J.M.; Gignoux, C.R.; Roth, L.A.; Kumar, R.; et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol 2014, 133, 670–678.e612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanowicz, D.; Hackett, T.-L.; Garmaroudi, F.S.; Günther, O.P.; Neumann, S.; Sutanto, E.N.; Ling, K.-M.; Kobor, M.S.; Kicic, A.; Stick, S.M.; et al. DNA methylation profiles of airway epithelial cells and pbmcs from healthy, atopic and asthmatic children. PLoS ONE 2012, 7, e44213. [Google Scholar] [CrossRef] [Green Version]
- Elgert, K.D. Immunology-Understanding the Immune System, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2009; p. 726. [Google Scholar]
- Alculumbre, S.; Pattarini, L. Purification of human dendritic cell subsets from peripheral blood. In Dendritic Cell Protocols; Segura, E., Onai, N., Eds.; Springer: New York, NY, USA, 2016; pp. 153–167. [Google Scholar]
- Brosseron, F.; Marcus, K.; May, C. Isolating peripheral lymphocytes by density gradient centrifugation and magnetic cell sorting. In Proteomic Profiling: Methods and Protocols; Posch, A., Ed.; Springer: New York, NY, USA, 2015; pp. 33–42. [Google Scholar]
- Gunawardhana, L.P.; Gibson, P.G.; Simpson, J.L.; Benton, M.C.; Lea, R.A.; Baines, K.J. Characteristic DNA methylation profiles in peripheral blood monocytes are associated with inflammatory phenotypes of asthma. Epigenetics 2014, 9, 1302–1316. [Google Scholar] [CrossRef] [Green Version]
- Wiencke, J.K.; Butler, R.; Hsuang, G.; Eliot, M.; Kim, S.; Sepulveda, M.A.; Siegel, D.; Houseman, E.A.; Kelsey, K.T. The DNA methylation profile of activated human natural killer cells. Epigenetics 2016, 11, 363–380. [Google Scholar] [CrossRef] [Green Version]
- Madore, A.-M.; Pain, L.; Boucher-Lafleur, A.-M.; Meloche, J.; Morin, A.; Simon, M.-M.; Ge, B.; Kwan, T.; Cheung, W.A.; Pastinen, T.; et al. Gsdma drives the most replicated association with asthma in naïve cd4+ t cells. bioRxiv 2019, 774760. [Google Scholar] [CrossRef] [Green Version]
- Morales-Nebreda, L.; McLafferty, F.S.; Singer, B.D. DNA methylation as a transcriptional regulator of the immune system. Transl. Res. 2019, 204, 1–18. [Google Scholar] [CrossRef]
- Farlik, M.; Halbritter, F.; Müller, F.; Choudry, F.A.; Ebert, P.; Klughammer, J.; Farrow, S.; Santoro, A.; Ciaurro, V.; Mathur, A.; et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 2016, 19, 808–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacoby, M.; Gohrbandt, S.; Clausse, V.; Brons, N.H.; Muller, C.P. Interindividual variability and co-regulation of DNA methylation differ among blood cell populations. Epigenetics 2012, 7, 1421–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laprise, C. The saguenay-lac-saint-jean asthma familial collection: The genetics of asthma in a young founder population. Genes Immun. 2014, 15, 247. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yergeau, D.; Wong, I.; Tuypens, T.; Tavernier, J.; Paul, C.; Baumann, M.; Auron, P.; Tenen, D.; Ackerman, S. Interleukin-5 receptor alpha subunit gene regulation in human eosinophil development: Identification of a unique cis-element that acts like an enhancer in regulating activity of the il-5r alpha promoter. Curr. Top. Microbiol. Immunobiol. 1996, 211, 173–187. [Google Scholar]
- Yang, I.V.; Pedersen, B.S.; Liu, A.H.; O’Connor, G.T.; Pillai, D.; Kattan, M.; Misiak, R.T.; Gruchalla, R.; Szefler, S.J.; Khurana Hershey, G.K.; et al. The nasal methylome and childhood atopic asthma. J. Allergy Clin. Immunol. 2017, 139, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Forno, E.; Wang, T.; Qi, C.; Yan, Q.; Xu, C.-J.; Boutaoui, N.; Han, Y.-Y.; Weeks, D.E.; Jiang, Y.; Rosser, F.; et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: A genome-wide study. Lancet Respir. Med. 2019, 7, 336–346. [Google Scholar] [CrossRef]
- Naranbhai, V.; Fairfax, B.P.; Makino, S.; Humburg, P.; Wong, D.; Ng, E.; Hill, A.V.S.; Knight, J.C. Genomic modulators of gene expression in human neutrophils. Nat. Commun. 2015, 6, 7545. [Google Scholar] [CrossRef] [Green Version]
- Kuramasu, A.; Saito, H.; Suzuki, S.; Watanabe, T.; Ohtsu, H. Mast cell-/basophil-specific transcriptional regulation of human l-histidine decarboxylase gene by cpg methylation in the promoter region. J. Biol. Chem. 1998, 273, 31607–31614. [Google Scholar] [CrossRef] [Green Version]
- Dunford, P.J.; Holgate, S.T. The role of histamine in asthma. In Histamine in Inflammation; Thurmond, R.L., Ed.; Springer: Boston, MA, USA, 2010; pp. 53–66. [Google Scholar]
- Hung, C.-H.; Wang, C.-C.; Suen, J.-L.; Sheu, C.-C.; Kuo, C.-H.; Liao, W.-T.; Yang, Y.-H.; Wu, C.-C.; Leung, S.-Y.; Lai, R.-S.; et al. Altered pattern of monocyte differentiation and monocyte-derived tgf-β1 in severe asthma. Sci. Rep. 2018, 8, 919. [Google Scholar] [CrossRef] [Green Version]
- Rivier, A.; Pène, J.; Rabesandratana, H.; Chanez, P.; Bousquet, J.; Campbell, A.M. Blood monocytes of untreated asthmatics exhibit some features of tissue macrophages. Clin. Exp. Immunol. 1995, 100, 314–318. [Google Scholar] [CrossRef]
- Zawada, A.M.; Schneider, J.S.; Michel, A.I.; Rogacev, K.S.; Hummel, B.; Krezdorn, N.; Müller, S.; Rotter, B.; Winter, P.; Obeid, R.; et al. DNA methylation profiling reveals differences in the 3 human monocyte subsets and identifies uremia to induce DNA methylation changes during differentiation. Epigenetics 2016, 11, 259–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekkers, K.F.; Neele, A.E.; Jukema, J.W.; Heijmans, B.T.; de Winther, M.P.J. Human monocyte-to-macrophage differentiation involves highly localized gain and loss of DNA methylation at transcription factor binding sites. Epigenet. Chrom. 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saradna, A.; Do, D.C.; Kumar, S.; Fu, Q.-L.; Gao, P. Macrophage polarization and allergic asthma. Transl. Res. 2018, 191, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Shahal, T.; Gabrieli, T.; Gilat, N.; Torchinsky, D.; Michaeli, Y.; Vogel, V.; Ebenstein, Y. Global modulation in DNA epigenetics during pro-inflammatory macrophage activation. Epigenetics 2019, 14, 1183–1193. [Google Scholar] [CrossRef]
- Wallner, S.; Schröder, C.; Leitão, E.; Berulava, T.; Haak, C.; Beißer, D.; Rahmann, S.; Richter, A.S.; Manke, T.; Bönisch, U.; et al. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenet. Chrom. 2016, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Puttur, F.; Gregory, L.G.; Lloyd, C.M. Airway macrophages as the guardians of tissue repair in the lung. Immunol. Cell Biol. 2019, 97, 246–257. [Google Scholar] [CrossRef]
- Van Helden, M.J.; Lambrecht, B.N. Dendritic cells in asthma. Curr. Opin. Immunol. 2013, 25, 745–754. [Google Scholar] [CrossRef]
- Zhang, X.; Ulm, A.; Somineni, H.K.; Oh, S.; Weirauch, M.T.; Zhang, H.-X.; Chen, X.; Lehn, M.A.; Janssen, E.M.; Ji, H. DNA methylation dynamics during ex vivo differentiation and maturation of human dendritic cells. Epigenet. Chrom. 2014, 7, 21. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Meng, L.; Zhang, Y. Epigenetic regulation of dendritic cell development and function. Cancer J. 2017, 23, 302–307. [Google Scholar] [CrossRef]
- Mishra, A. Metabolic plasticity in dendritic cell responses: Implications in allergic asthma. J. Immunol. Res. 2017, 2017, 5134760. [Google Scholar] [CrossRef] [Green Version]
- Fedulov, A.V.; Kobzik, L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am. J. Respir. Cell Mol. Biol 2011, 44, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, D.J.; Kobzik, L.; Yang, Z.; McGuire, C.C.; Fedulov, A.V. Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy. Am. J. Phys. Lung Cell. Mol. Phys. 2017, 313, L395–L405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidl, C.; Delacher, M.; Huehn, J.; Feuerer, M. Epigenetic mechanisms regulating t-cell responses. J. Allergy Clin. Immunol. 2018, 142, 728–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, M.L.; Ellis, A.K. The role of epigenetics in the developmental origins of allergic disease. Ann. Allergy Asthma Immunol. 2011, 106, 355–361. [Google Scholar] [CrossRef]
- Tsagaratou, A.; Äijö, T.; Lio, C.-W.J.; Yue, X.; Huang, Y.; Jacobsen, S.E.; Lähdesmäki, H.; Rao, A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in t-cell development and differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E3306–E3315. [Google Scholar] [CrossRef] [Green Version]
- Lal, G.; Zhang, N.; van der Touw, W.; Ding, Y.; Ju, W.; Bottinger, E.P.; Reid, S.P.; Levy, D.E.; Bromberg, J.S. Epigenetic regulation of foxp3 expression in regulatory t cells by DNA methylation. J. Immunol. 2009, 182, 259–273. [Google Scholar] [CrossRef]
- Huehn, J.; Polansky, J.K.; Hamann, A. Epigenetic control of foxp3 expression: The key to a stable regulatory t-cell lineage? Nat. Rev. Immunol. 2009, 9, 83. [Google Scholar] [CrossRef]
- Toker, A.; Engelbert, D.; Garg, G.; Polansky, J.K.; Floess, S.; Miyao, T.; Baron, U.; Düber, S.; Geffers, R.; Giehr, P.; et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 2013, 190, 3180. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-P.; Leonard, W.J. Creb/atf-dependent t cell receptor-induced foxp3 gene expression: A role for DNA methylation. J. Exp. Med. 2007, 204, 1543–1551. [Google Scholar] [CrossRef]
- Lawless, O.J.; Bellanti, J.A.; Brown, M.L.; Sandberg, K.; Umans, J.G.; Zhou, L.; Chen, W.; Wang, J.; Wang, K.; Zheng, S.G. In vitro induction of t regulatory cells by a methylated cpg DNA sequence in humans: Potential therapeutic applications in allergic and autoimmune diseases. Allergy Asthma Proc. 2018, 39, 143–152. [Google Scholar] [CrossRef]
- Yu, C.-X.; Bai, L.-Y.; Lin, J.-J.; Li, S.-B.; Chen, J.-Y.; He, W.-J.; Yu, X.-M.; Cui, X.-P.; Wang, H.-L.; Chen, Y.-Z.; et al. Rhpld2 inhibits airway inflammation in an asthmatic murine model through induction of stable cd25+ foxp3+ tregs. Mol. Immunol. 2018, 101, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.; McDonald-Hyman, C.; Noth, E.M.; Pratt, B.; Hammond, S.K.; Balmes, J.; Tager, I. Ambient air pollution impairs regulatory t-cell function in asthma. J. Allergy Clin. Immunol. 2010, 126, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Scharer, C.D.; Barwick, B.G.; Youngblood, B.A.; Ahmed, R.; Boss, J.M. Global DNA methylation remodeling accompanies cd8 t cell effector function. J. Immunol. 2013, 191, 3419–3429. [Google Scholar] [CrossRef] [Green Version]
- Youngblood, B.; Hale, J.S.; Kissick, H.T.; Ahn, E.; Xu, X.; Wieland, A.; Araki, K.; West, E.E.; Ghoneim, H.E.; Fan, Y.; et al. Effector cd8 t cells dedifferentiate into long-lived memory cells. Nature 2017, 552, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Shaknovich, R.; Cerchietti, L.; Tsikitas, L.; Kormaksson, M.; De, S.; Figueroa, M.E.; Ballon, G.; Yang, S.N.; Weinhold, N.; Reimers, M.; et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center b-cell differentiation. Blood 2011, 118, 3559–3569. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, P.M.; Teater, M.; Chambwe, N.; Kormaksson, M.; Redmond, D.; Ishii, J.; Vuong, B.; Chaudhuri, J.; Melnick, A.; Vasanthakumar, A.; et al. DNA methylation dynamics of germinal center b cells are mediated by aid. Cell Rep. 2015, 12, 2086–2098. [Google Scholar] [CrossRef] [Green Version]
- Pascual, M.; Suzuki, M.; Isidoro-Garcia, M.; Padrón, J.; Turner, T.; Lorente, F.; Dávila, I.; Greally, J.M. Epigenetic changes in b lymphocytes associated with house dust mite allergic asthma. Epigenetics 2011, 6, 1131–1137. [Google Scholar] [CrossRef]
- Kim, J.H.; Jang, Y.J. Role of natural killer cells in airway inflammation. Allergy Asthma Immunol. Res. 2018, 10, 448–456. [Google Scholar] [CrossRef]
- Duvall, M.G.; Barnig, C.; Cernadas, M.; Ricklefs, I.; Krishnamoorthy, N.; Grossman, N.L.; Bhakta, N.R.; Fahy, J.V.; Bleecker, E.R.; Castro, M.; et al. Natural killer cell-mediated inflammation resolution is disabled in severe asthma. Sci. Immunol. 2017, 2, eaam5446. [Google Scholar] [CrossRef] [Green Version]
- Gorska, M.M. Natural killer cells in asthma. Curr. Opin. Allergy Clin. Immunol. 2017, 17, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Mathias, C.B. Natural killer cells in the development of asthma. Curr. Allergy Asthma Rep. 2014, 15, 500. [Google Scholar] [CrossRef] [PubMed]
- Vieira-de-Abreu, A.; Campbell, R.A.; Weyrich, A.S.; Zimmerman, G.A. Platelets: Versatile effector cells in hemostasis, inflammation, and the immune continuum. Semin. Immunopathol. 2012, 34, 5–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Casado, C.; Villaseñor, A.; Rodriguez-Nogales, A.; Bueno, J.L.; Barber, D.; Escribese, M.M. Understanding platelets in infectious and allergic lung diseases. Int. J. Mol. Sci. 2019, 20, 1730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambrecht, B.N.; Hammad, H.; Fahy, J.V. The cytokines of asthma. Immunity 2019, 50, 975–991. [Google Scholar] [CrossRef]
- Mims, J.W. Asthma: Definitions and pathophysiology. Int. Forum Allergy Rhinol. 2015, 5, S2–S6. [Google Scholar] [CrossRef]
- McErlean, P.; Favoreto, S.; Costa, F.F.; Shen, J.; Quraishi, J.; Biyasheva, A.; Cooper, J.J.; Scholtens, D.M.; Vanin, E.F.; de Bonaldo, M.F.; et al. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Med. Genomics 2014, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Larouche, M.; Gagné-Ouellet, V.; Boucher-Lafleur, A.-M.; Larose, M.-C.; Plante, S.; Madore, A.-M.; Laviolette, M.; Flamand, N.; Chakir, J.; Laprise, C. Methylation profiles of il33 and ccl26 in bronchial epithelial cells are associated with asthma. Epigenomics 2018, 10, 1555–1568. [Google Scholar] [CrossRef]
- Nicodemus-Johnson, J.; Naughton, K.A.; Sudi, J.; Hogarth, K.; Naurekas, E.T.; Nicolae, D.L.; Sperling, A.I.; Solway, J.; White, S.R.; Ober, C. Genome-wide methylation study identifies an il-13-induced epigenetic signature in asthmatic airways. Am. J. Respir. Crit. Care Med. 2016, 193, 376–385. [Google Scholar] [CrossRef] [Green Version]
- Ooi, A.T.; Ram, S.; Kuo, A.; Gilbert, J.L.; Yan, W.; Pellegrini, M.; Nickerson, D.W.; Chatila, T.A.; Gomperts, B.N. Identification of an interleukin 13-induced epigenetic signature in allergic airway inflammation. Am. J. Transl. Res. 2012, 4, 219–228. [Google Scholar]
- Scantamburlo, G.; Vanoni, S.; Dossena, S.; Soyal, S.M.; Bernardinelli, E.; Civello, D.A.; Patsch, W.; Paulmichl, M.; Nofziger, C. Interleukin-4 induces cpg site-specific demethylation of the pendrin promoter in primary human bronchial epithelial cells. Cell. Phys. Biochem. 2017, 41, 1491–1502. [Google Scholar] [CrossRef]
- Vanoni, S.; Scantamburlo, G.; Dossena, S.; Paulmichl, M.; Nofziger, C. Interleukin-mediated pendrin transcriptional regulation in airway and esophageal epithelia. Int. J. Mol. Sci. 2019, 20, 731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baccarelli, A.; Rusconi, F.; Bollati, V.; Catelan, D.; Accetta, G.; Hou, L.; Barbone, F.; Bertazzi, P.A.; Biggeri, A. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics 2012, 4, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, C.; Biagini Myers, J.M.; Ji, H.; Metz, K.; Martin, L.J.; Lindsey, M.; He, H.; Powers, R.; Ulm, A.; Ruff, B.; et al. Vanin-1 expression and methylation discriminate pediatric asthma corticosteroid treatment response. J. Allergy Clin. Immunol. 2015, 136, 923–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Biagini Myers, J.M.; Yadagiri, V.K.; Ulm, A.; Chen, X.; Weirauch, M.T.; Khurana Hershey, G.K.; Ji, H. Nasal DNA methylation differentiates corticosteroid treatment response in pediatric asthma: A pilot study. PLoS ONE 2017, 12, e0186150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salter, B.; Pray, C.; Radford, K.; Martin, J.G.; Nair, P. Regulation of human airway smooth muscle cell migration and relevance to asthma. Respir. Res. 2017, 18, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, M.M.; Lavender, P.; Kuo, C.-H.S.; Galea, F.; Michaeloudes, C.; Flanagan, J.M.; Fan Chung, K.; Adcock, I.M. DNA methylation modules in airway smooth muscle are associated with asthma severity. Eur. Respir. J. 2018, 51, 1701068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allum, F.; Shao, X.; Guénard, F.; Simon, M.-M.; Busche, S.; Caron, M.; Lambourne, J.; Lessard, J.; Tandre, K.; Hedman, Å.K.; et al. Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat. Commun. 2015, 6, 7211. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.W. Activation states of blood eosinophils in asthma. Clin. Exp. Allergy 2014, 44, 482–498. [Google Scholar] [CrossRef] [Green Version]
- Draijer, C.; Peters-Golden, M. Alveolar macrophages in allergic asthma: The forgotten cell awakes. Curr. Allergy Asthma Rep. 2017, 17, 12. [Google Scholar] [CrossRef] [Green Version]
- Wen, T.; Rothenberg, M.E. The regulatory function of eosinophils. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Loxham, M.; Davies, D.E. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J. Allergy Clin. Immunol. 2017, 139, 1736–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, K.; Isogai, S.; Tamaoka, M.; Inase, N.; Akashi, T.; Martin, J.G.; Yoshizawa, Y. Depletion of cd8+ t cells enhances airway remodelling in a rodent model of asthma. Immunology 2009, 126, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Torrone, D.; Kuriakose, J.; Moors, K.; Jiang, H.; Niedzwiecki, M.; Perera, F.; Miller, R. Reproducibility and intraindividual variation over days in buccal cell DNA methylation of two asthma genes, interferon γ (ifnγ) and inducible nitric oxide synthase (inos). Clin. Epigenet. 2012, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.H.; Lovinsky-Desir, S.; Yan, B.; Torrone, D.; Lawrence, J.; Jezioro, J.R.; Perzanowski, M.; Perera, F.P.; Chillrud, S.N.; Miller, R.L. Effect of personal exposure to black carbon on changes in allergic asthma gene methylation measured 5 days later in urban children: Importance of allergic sensitization. Clin. Epigenet. 2017, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breton, C.V.; Byun, H.-M.; Wang, X.; Salam, M.T.; Siegmund, K.; Gilliland, F.D. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am. J. Respir. Crit. Care Med. 2011, 184, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brugha, R.; Lowe, R.; Henderson, A.J.; Holloway, J.W.; Rakyan, V.; Wozniak, E.; Mahmud, N.; Seymour, K.; Grigg, J.; Shaheen, S.O. DNA methylation profiles between airway epithelium and proxy tissues in children. Acta Paediatr. 2017, 106, 2011–2016. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.D.; O’Byrne, P.M. Biologics and the lung: Tslp and other epithelial cell-derived cytokines in asthma. Pharmacol. Ther. 2017, 169, 104–112. [Google Scholar] [CrossRef]
- Xu, M.-H.; Yuan, F.-L.; Wang, S.-J.; Xu, H.-Y.; Li, C.-W.; Tong, X. Association of interleukin-18 and asthma. Inflammation 2017, 40, 324–327. [Google Scholar] [CrossRef]
- Koch, S.; Sopel, N.; Finotto, S. Th9 and other il-9-producing cells in allergic asthma. Semin. Immunopathol. 2017, 39, 55–68. [Google Scholar] [CrossRef]
- Walsh, G.M. Biologics targeting il-5, il-4 or il-13 for the treatment of asthma—An update. Expert Rev. Clin. Immunol. 2017, 13, 143–149. [Google Scholar] [CrossRef]
- Bonser, L.R.; Erle, D.J. Chapter one—The airway epithelium in asthma. In Advances in Immunology; Alt, F., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 142, pp. 1–34. [Google Scholar]
- Bartemes, K.R.; Kita, H. Dynamic role of epithelium-derived cytokines in asthma. Clin. Immunol. 2012, 143, 222–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, K.D.; Uzvolgyi, E.; Liang, G.; Talmadge, C.; Sumegi, J.; Gonzales, F.A.; Jones, P.A. The human DNA methyltransferases (dnmts) 1, 3a and 3b: Coordinate mrna expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999, 27, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamper, C.J.; Agoston, A.T.; Nelson, W.G.; Powell, J.D. Identification of DNA methyltransferase 3a as a t cell receptor-induced regulator of th1 and th2 differentiation. J. Immunol. 2009, 183, 2267–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervouet, E.; Peixoto, P.; Delage-Mourroux, R.; Boyer-Guittaut, M.; Cartron, P.-F. Specific or not specific recruitment of dnmts for DNA methylation, an epigenetic dilemma. Clin. Epigenet. 2018, 10, 17. [Google Scholar] [CrossRef]
- Trian, T.; Allard, B.; Ozier, A.; Maurat, E.; Dupin, I.; Thumerel, M.; Ousova, O.; Gillibert-Duplantier, J.; Le Morvan, V.; Begueret, H.; et al. Selective dysfunction of p53 for mitochondrial biogenesis induces cellular proliferation in bronchial smooth muscle from asthmatic patients. J. Allergy Clin. Immunol. 2016, 137, 1717–1726.e1713. [Google Scholar] [CrossRef]
- Kubo, T.; Tsujiwaki, M.; Hirohashi, Y.; Tsukahara, T.; Kanaseki, T.; Nakatsugawa, M.; Hasegawa, T.; Torigoe, T. Differential bronchial epithelial response regulated by δnp63: A functional understanding of the epithelial shedding found in asthma. Lab. Investig. 2019, 99, 158–168. [Google Scholar] [CrossRef]
- Xu, L.; Sun, W.J.; Jia, A.J.; Qiu, L.L.; Xiao, B.; Mu, L.; Li, J.M.; Zhang, X.F.; Wei, Y.; Peng, C.; et al. Mbd2 regulates differentiation and function of th17 cells in neutrophils-dominant asthma via hif-1α. J. Inflamm. 2018, 15. [Google Scholar] [CrossRef]
- Sun, W.; Xiao, B.; Jia, A.; Qiu, L.; Zeng, Q.; Liu, D.; Yuan, Y.; Jia, J.; Zhang, X.; Xiang, X. Mbd2-mediated th17 differentiation in severe asthma is associated with impaired socs3 expression. Exp. Cell Res. 2018, 371, 196–204. [Google Scholar] [CrossRef]
- Lokk, K.; Modhukur, V.; Rajashekar, B.; Märtens, K.; Mägi, R.; Kolde, R.; Koltšina, M.; Nilsson, T.K.; Vilo, J.; Salumets, A.; et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014, 15, r54. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Zhou, Y.; Wang, C.-M.; Huang, T.H.M.; Jin, V.X. Integration of DNA methylation and gene transcription across nineteen cell types reveals cell type-specific and genomic region-dependent regulatory patterns. Sci. Rep. 2017, 7, 3626. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.D.; Jones, M.J.; Meaney, M.J.; Turecki, G.; Kobor, M.S. Becon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatr. 2017, 7, e1187. [Google Scholar] [CrossRef] [Green Version]
- Acevedo, N.; Reinius, L.E.; Greco, D.; Gref, A.; Orsmark-Pietras, C.; Persson, H.; Pershagen, G.; Hedlin, G.; Melén, E.; Scheynius, A.; et al. Risk of childhood asthma is associated with cpg-site polymorphisms, regional DNA methylation and mrna levels at the gsdmb/ormdl3 locus. Hum. Mol. Gen. 2015, 24, 875–890. [Google Scholar] [CrossRef]
- Ewing, E.; Kular, L.; Fernandes, S.J.; Karathanasis, N.; Lagani, V.; Ruhrmann, S.; Tsamardinos, I.; Tegner, J.; Piehl, F.; Gomez-Cabrero, D.; et al. Combining evidence from four immune cell types identifies DNA methylation patterns that implicate functionally distinct pathways during multiple sclerosis progression. EBioMedicine 2019, 43, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roadmap Epigenomics, C.; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317. [Google Scholar]
- Yao, Y.; Welp, T.; Liu, Q.; Niu, N.; Wang, X.; Britto, C.J.; Krishnaswamy, S.; Chupp, G.L.; Montgomery, R.R. Multiparameter single cell profiling of airway inflammatory cells. Cytom. B Clin. Cytom. 2017, 92, 12–20. [Google Scholar] [CrossRef]
- Farlik, M.; Sheffield, N.C.; Nuzzo, A.; Datlinger, P.; Schönegger, A.; Klughammer, J.; Bock, C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015, 10, 1386–1397. [Google Scholar] [CrossRef] [Green Version]
- Bohrson, C.L.; Barton, A.R.; Lodato, M.A.; Rodin, R.E.; Luquette, L.J.; Viswanadham, V.V.; Gulhan, D.C.; Cortés-Ciriano, I.; Sherman, M.A.; Kwon, M.; et al. Linked-read analysis identifies mutations in single-cell DNA-sequencing data. Nat. Gen. 2019, 51, 749–754. [Google Scholar] [CrossRef]





| Cytokines | AECs | Eosinophils |
|---|---|---|
| IL-1 | ACP5, CCL26, DAB2IP, FBXW11, HNMT, PXDN, RIPK2, TRIP6, SUCNR1 | ACP5, FOXP1 |
| IL-2 | ACP5, HNMT, LAG3, PTPRC, RIPK2, STAT5A, ZFP36 | ACP5 |
| IL-3 | NFIL3 | PRKCZ |
| IL-4 | EPX, NFIL3 | EPX, IL4, PRKCZ, ZFPM1 |
| IL-5 | EPX, IL5RA | EPX, IL1RL1, IL5RA, PRKCZ |
| IL-6 | SORL1 | |
| IL-7 | STAT5A | |
| IL-8 | PRG3, PTPRC, TNFSF12 | CAMP, PLA2G1B, PRG3 |
| IL-9 | STAT5A | |
| IL-10 | EPX | EPX, PRKCZ, TRIB2 |
| IL-12 | RIPK2 | FOXP1 |
| IL-13 | ALOX15, IL13 | IL4 |
| IL-14 | METRNL | |
| IL-15 | STAT5A | |
| IL-16 | RIPK2 | |
| IL-18 | RIPK2 | |
| IL-21 | FOXP1 | |
| IL-33 | IL1RL1 | |
| Interferon (INF) | EVL, IFNGR2, LRRFIP1, MED1, TANK | EVL |
| Transforming growth factor (TGF) | ADAMTSL2, FERMT2, GCNT2, IL13, PTK2, SKI | ENG, GDF5 |
| Tumor necrosis factor (TNF) | CCL26, DICER1, MAP3K4, PTPRC, ZFP36 | DICER1, FOXP1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudon Thibeault, A.-A.; Laprise, C. Cell-Specific DNA Methylation Signatures in Asthma. Genes 2019, 10, 932. https://doi.org/10.3390/genes10110932
Hudon Thibeault A-A, Laprise C. Cell-Specific DNA Methylation Signatures in Asthma. Genes. 2019; 10(11):932. https://doi.org/10.3390/genes10110932
Chicago/Turabian StyleHudon Thibeault, Andrée-Anne, and Catherine Laprise. 2019. "Cell-Specific DNA Methylation Signatures in Asthma" Genes 10, no. 11: 932. https://doi.org/10.3390/genes10110932
APA StyleHudon Thibeault, A.-A., & Laprise, C. (2019). Cell-Specific DNA Methylation Signatures in Asthma. Genes, 10(11), 932. https://doi.org/10.3390/genes10110932




