Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription
Abstract
:1. Introduction: Germline, Soma and Embryonic Stem Cells
2. Differentiation of Germ Cells
3. Germ Cell Differentiation Factors
3.1. Maternally Contributed Factors
3.2. Diffusible Signaling Molecules
3.3. Germ-Cell-Specific Transcription Factors
3.4. Polycomb Repressive Complexes
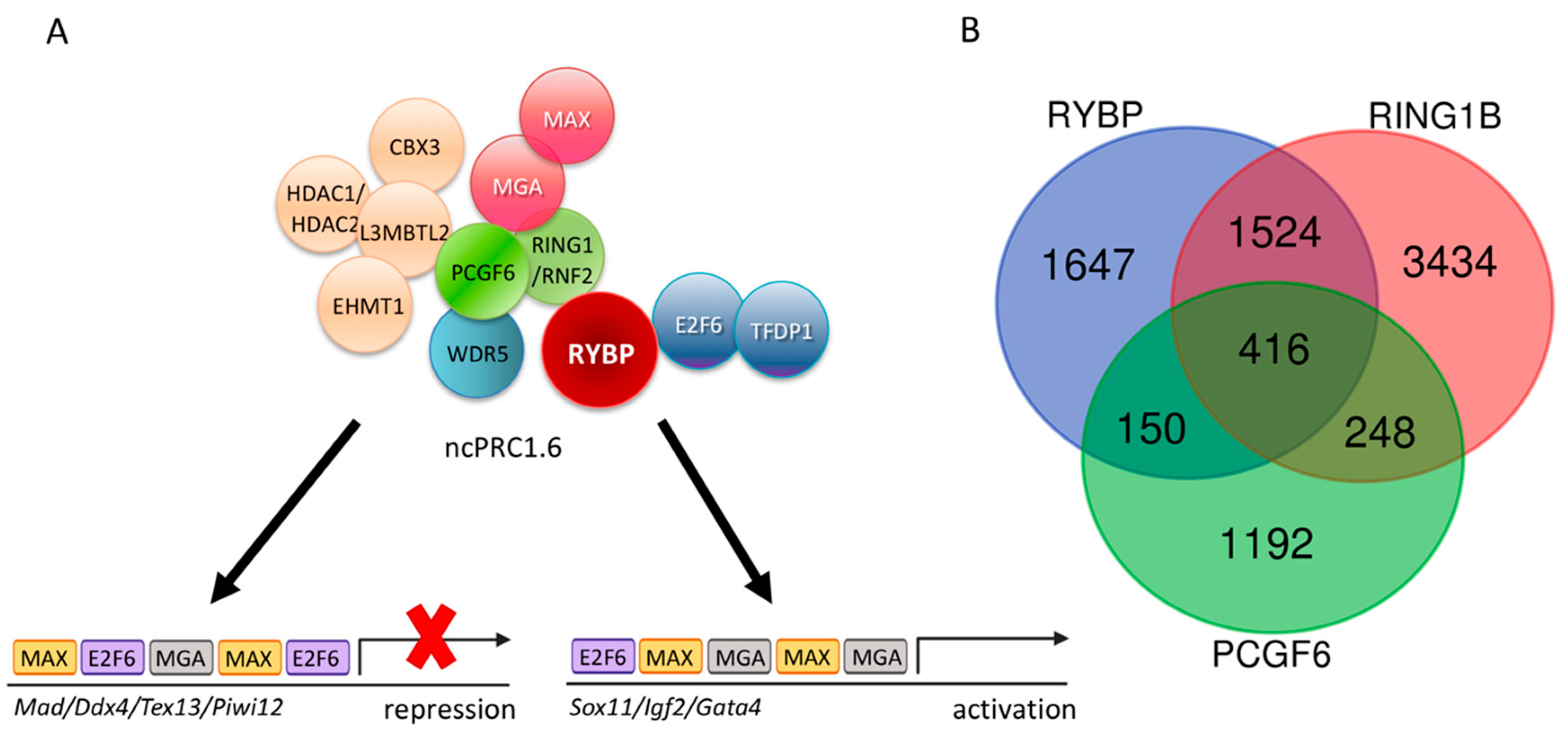
Non-canonical PRC1.6, A Major Repressor of Germ Cell Fate
4. Interactions of BMP and RA Signalization Pathways and PRC Complexes in Germ-Cell-Specific Regulatory Processes
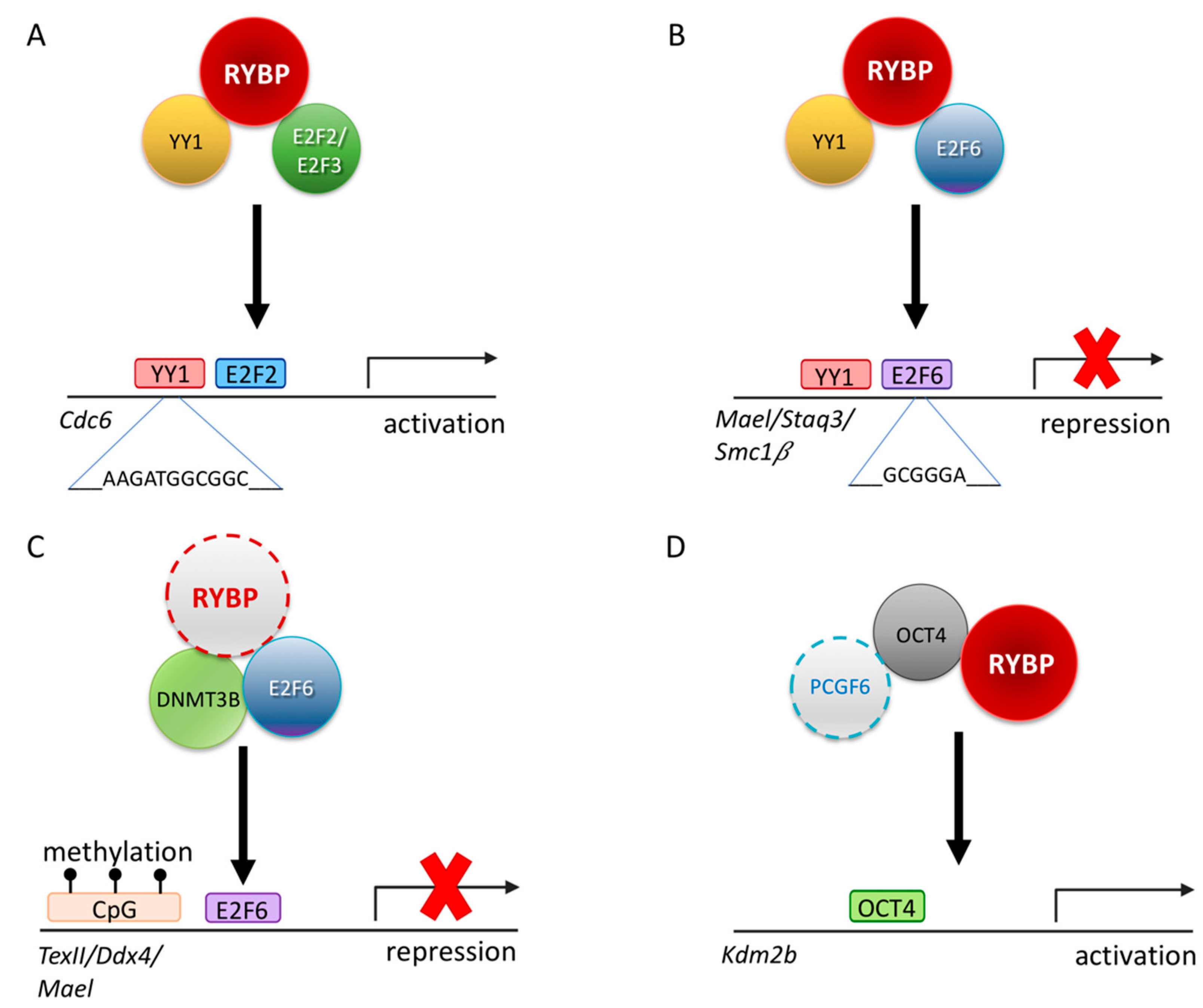
5. Connections between RYBP and Germ Cell Fate Regulation
6. Commonalities between Germ Cells and ES Cells
7. RYBP Is Connected to Germline Specific Functions by a Plethora of its Binding Partners
7.1. Natively Unfolded Structure of the RYBP Enables It to Affect Germ-Cell-Specific Functions in Multiple Ways
7.2. RYBP is Connected to Germ-Cell-Specific Apoptotic Processes via FANK1
7.3. RYBP Can Contribute to the Regulation of Germ-Cell-Specific Gene Expression Directly as a Subunit of ncPRC1.6
7.4. RYBP Regulates Germ-Cell-Specific Gene Expression Indirectly through its Multiple Connection with the Ubiquitin System
7.5. RYBP Directly Interacts with OCT4/POU5F1 and Can Be Targeted to Pluripotency Target Genes Bound by OCT4/POU5F1
7.6. Loss of Rybp Affects the RA Signalization Pathway and the Level of Nanog Simultaneously in ES Cells
7.7. RYBP and DDX5 is Connected to Reprogramming via a microrna Based Regulatory Loop
8. Methods
8.1. ChIP-seq Analysis
8.2. RNA-seq Analysis
9. Closing Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wylie, C. Germ cells. Cell 1999, 96, 165–174. [Google Scholar] [CrossRef]
- Seydoux, G.; Braun, R.E. Pathway to Totipotency: Lessons from Germ Cells. Cell 2006, 127, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Leitch, H.G.; Smith, A. The mammalian germline as a pluripotency cycle. Development 2013, 140, 2495–2501. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, J.W.; Koster, J.; Lodder, P.; Repping, S.; Hamer, G. Massive expression of germ cell-specific genes is a hallmark of cancer and a potential target for novel treatment development. Oncogene 2018, 37, 5694–5700. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Saitou, M. Germ cell specification in mice. Curr. Opin. Genet. Dev. 2009, 19, 386–395. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931–942. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Primordial Germ Cells in Mice. Cold Spring Harb. Perspect. Biol. 2012, 4, a008375. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, E.; Dietmann, S.; Murakami, K.; Günesdogan, U.; Tang, F.; Bao, S.; Diamanti, E.; Lao, K.; Gottgens, B.; Azim Surani, M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013, 15, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Jostes, S.; Schorle, H. Signals and transcription factors for specification of human germ cells. Stem Cell Investig. 2018, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Tang, W.W.C.; Azim Surani, M. Germ cell specification and pluripotency in mammals: A perspective from early embryogenesis. Reprod. Med. Biol. 2014, 13, 203–215. [Google Scholar] [CrossRef]
- Magnusdottir, E.; Surani, M.A. How to make a primordial germ cell. Development 2014, 141, 245–252. [Google Scholar] [CrossRef]
- Wang, X.; Liao, T.; Wan, C.; Yang, X.; Zhao, J.; Fu, R.; Yao, Z.; Huang, Y.; Shi, Y.; Chang, G.; et al. Efficient generation of human primordial germ cell-like cells from pluripotent stem cells in a methylcellulose-based 3D system at large scale. PeerJ 2019, 6, e6143. [Google Scholar] [CrossRef]
- Mitsunaga, S.; Shioda, K.; Isselbacher, K.J.; Hanna, J.H.; Shioda, T. Generation of Human Primordial Germ Cell-like Cells at the Surface of Embryoid Bodies from Primed-pluripotency Induced Pluripotent Stem Cells. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Nagamatsu, G.; Suda, T. Conversion of Primordial Germ Cells to Pluripotent Stem Cells: Methods for Cell Tracking and Culture Conditions. Method. Mol. Biol. 2013, 1052, 49–56. [Google Scholar]
- Surani, M.A.; Durcova-Hills, G.; Hajkova, P.; Hayashi, K.; Tee, W.W. Germ Line, Stem Cells, and Epigenetic Reprogramming. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 9–15. [Google Scholar] [CrossRef]
- Wei, Y.; Schatten, H.; Sun, Q.-Y. Environmental epigenetic inheritance through gametes and implications for human reproduction. Hum. Reprod. Update 2015, 21, 194–208. [Google Scholar] [CrossRef]
- Guéant, J.-L.; Chéry, C.; Oussalah, A.; Nadaf, J.; Coelho, D.; Josse, T.; Flayac, J.; Robert, A.; Koscinski, I.; Gastin, I.; et al. A PRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat. Commun. 2018, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Horsthemke, B. A critical view on transgenerational epigenetic inheritance in humans. Nat. Commun. 2018, 9, 2973. [Google Scholar] [CrossRef] [PubMed]
- Cinalli, R.M.; Rangan, P.; Lehmann, R. Germ Cells Are Forever. Cell 2008, 132, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Extavour, C.G.; Akam, M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development 2003, 130, 5869–5884. [Google Scholar] [CrossRef] [PubMed]
- Seervai, R.N.H.; Wessel, G.M. Lessons for inductive germline determination. Mol. Reprod. Dev. 2013, 80, 590–609. [Google Scholar] [CrossRef]
- Krishnakumar, P.; Dosch, R. Germ Cell Specification: The Evolution of a Recipe to Make Germ Cells. In Germ Cell; Ahmed, R.G., Ed.; InTech Open: London, UK, 2018. [Google Scholar]
- Kumano, G. Evolution of germline segregation processes in animal development. Dev. Growth Differ. 2015, 57, 324–332. [Google Scholar] [CrossRef]
- Voronina, E.; Seydoux, G.; Sassone-Corsi, P.; Nagamori, I. RNA Granules in Germ Cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002774. [Google Scholar] [CrossRef]
- Strome, S.; Lehmann, R. Germ Versus Soma Decisions: Lessons from Flies and Worms. Science 2007, 316, 392–393. [Google Scholar] [CrossRef]
- Weismann, A. Das Keimplasma; Auflage, Ed.; Fischer: Jena, Germany, 1892. [Google Scholar]
- Li, L.; Zheng, P.; Dean, J. Maternal control of early mouse development. Development 2010, 137, 859–870. [Google Scholar] [CrossRef]
- Carter, M.G.; Hamatani, T.; Sharov, A.A.; Carmack, C.E.; Qian, Y.; Aiba, K.; Ko, N.T.; Dudekula, D.B.; Brzoska, P.M.; Hwang, S.S.; et al. In Situ-Synthesized Novel Microarray Optimized for Mouse Stem Cell and Early Developmental Expression Profiling. Genom. Res. 2003, 13, 1011–1021. [Google Scholar] [CrossRef]
- Wang, Q.T.; Piotrowska, K.; Ciemerych, M.A.; Milenkovic, L.; Scott, M.P.; Davis, R.W.; Zernicka-Goetz, M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 2004, 6, 133–144. [Google Scholar] [CrossRef]
- Takaoka, K.; Hamada, H. Cell fate decisions and axis determination in the early mouse embryo. Development 2012, 139, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsburg, M.; Snow, M.H.; McLaren, A. Primordial germ cells in the mouse embryo during gastrulation. Development 1990, 110, 521–528. [Google Scholar]
- Anderson, R.; Copeland, T.K.; Schöler, H.; Heasman, J.; Wylie, C. The onset of germ cell migration in the mouse embryo. Mech. Dev. 2000, 91, 61–68. [Google Scholar] [CrossRef]
- Spiegelman, M.; Bennett, D. A light- and electron-microscopic study of primordial germ cells in the early mouse embryo. J. Embryol. Exp. Morphol. 1973, 30, 97–118. [Google Scholar]
- Clark, J.M.; Eddy, E.M. Fine structural observations on the origin and associations of primordial germ cells of the mouse. Dev. Biol. 1975, 47, 136–155. [Google Scholar] [CrossRef]
- Spiller, C.; Wilhelm, D.; Koopman, P. Cell cycle analysis of fetal germ cells during sex differentiation in mice. Biol. Cell 2009, 101, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Ewen, K.A.; Koopman, P. Mouse germ cell development: From specification to sex determination. Mol. Cell. Endocrinol. 2010, 323, 76–93. [Google Scholar] [CrossRef]
- Kotaja, N.; Sassone-Corsi, P. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007, 8, 85–90. [Google Scholar] [CrossRef]
- Juliano, C.E.; Swartz, S.Z.; Wessel, G.M. A conserved germline multipotency program. Development 2010, 137, 4113–4126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha-da-Silva, L.; Armelin-Correa, L.; Cantão, I.H.; Flister, V.J.F.; Nunes, M.; Stumpp, T. Expression of genome defence protein members in proliferating and quiescent rat male germ cells and the Nuage dynamics. PLoS ONE 2019, 14, e0217941. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Takamatsu, K.; Chuma, S.; Kojima-Kita, K.; Shiromoto, Y.; Asada, N.; Toyoda, A.; Fujiyama, A.; et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010, 24, 887–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soper, S.F.C.; van der Heijden, G.W.; Hardiman, T.C.; Goodheart, M.; Martin, S.L.; de Boer, P.; Bortvin, A. Mouse Maelstrom, a Component of Nuage, Is Essential for Spermatogenesis and Transposon Repression in Meiosis. Dev. Cell 2008, 15, 285–297. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.J.; Crossan, G.P. DNA cross-link repair safeguards genomic stability during premeiotic germ cell development. Nat. Genet. 2019, 51, 1283–1294. [Google Scholar] [CrossRef]
- Illmensee, K.; Mahowald, A.P. Transplantation of Posterior Polar Plasm in Drosophila. Induction of Germ Cells at the Anterior Pole of the Egg. Proc. Natl. Acad. Sci. USA 1974, 71, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Houston, D.W.; King, M.L. Germ plasm and molecular determinants of germ cell fate. Curr. Top. Dev. Biol. 2000, 50, 155–181. [Google Scholar]
- Lawson, K.A.; Dunn, N.R.; Roelen, B.A.J.; Zeinstra, L.M.; Davis, A.M.; Wright, C.V.E.; Korving, J.P.W.F.M.; Hogan, B.L.M. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999, 13, 424–436. [Google Scholar] [CrossRef]
- Saitou, M.; Barton, S.C.; Surani, M.A. A molecular programme for the specification of germ cell fate in mice. Nature 2002, 418, 293–300. [Google Scholar] [CrossRef]
- Bowles, J. Retinoid Signaling Determines Germ Cell Fate in Mice. Science 2006, 312, 596–600. [Google Scholar] [CrossRef] [Green Version]
- Yamashiro, C.; Hirota, T.; Kurimoto, K.; Nakamura, T.; Yabuta, Y.; Nagaoka, S.I.; Ohta, H.; Yamamoto, T.; Saitou, M. Persistent Requirement and Alteration of the Key Targets of PRDM1 During Primordial Germ Cell Development in Mice1. Biol. Reprod. 2016, 94, 7. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Wernig, M.; Jaenisch, R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007, 25, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Silva, J.; Colby, D.; Nichols, J.; Nijmeijer, B.; Robertson, M.; Vrana, J.; Jones, K.; Grotewold, L.; Smith, A. Nanog safeguards pluripotency and mediates germline development. Nature 2007, 450, 1230–1234. [Google Scholar] [CrossRef] [PubMed]
- Kalmar, T.; Lim, C.; Hayward, P.; Muñoz-Descalzo, S.; Nichols, J.; Garcia-Ojalvo, J.; Martinez Arias, A. Regulated Fluctuations in Nanog Expression Mediate Cell Fate Decisions in Embryonic Stem Cells. PLoS Biol. 2009, 7, e1000149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Hoei-Hansen, C.E.; Almstrup, K.; Nielsen, J.E.; Brask Sonne, S.; Graem, N.; Skakkebaek, N.E.; Leffers, H.; Rajpert-De Meyts, E. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology 2005, 47, 48–56. [Google Scholar] [CrossRef]
- Murakami, K.; Günesdogan, U.; Zylicz, J.J.; Tang, W.W.C.; Sengupta, R.; Kobayashi, T.; Kim, S.; Butler, R.; Dietmann, S.; Surani, M.A. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature 2016, 529, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Sowpati, D.T.; Ramamoorthy, S.; Mishra, R.K. Expansion of the polycomb system and evolution of complexity. Mech. Dev. 2015, 138, 97–112. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Stock, J.K.; Giadrossi, S.; Casanova, M.; Brookes, E.; Vidal, M.; Koseki, H.; Brockdorff, N.; Fisher, A.G.; Pombo, A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007, 9, 1428–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brookes, E.; Pombo, A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009, 10, 1213–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Pirrotta, V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef]
- Maezawa, S.; Hasegawa, K.; Yukawa, M.; Sakashita, A.; Alavattam, K.G.; Andreassen, P.R.; Vidal, M.; Koseki, H.; Barski, A.; Namekawa, S.H. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017, 31, 1693–1703. [Google Scholar] [CrossRef] [Green Version]
- Yokobayashi, S.; Liang, C.-Y.; Kohler, H.; Nestorov, P.; Liu, Z.; Vidal, M.; van Lohuizen, M.; Roloff, T.C.; Peters, A.H.F.M. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 2013, 495, 236–240. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Lai, P.; Jia, J.; Song, Y.; Xia, Q.; Huang, K.; He, N.; Ping, W.; Chen, J.; Yang, Z.; et al. RNA Helicase DDX5 Inhibits Reprogramming to Pluripotency by miRNA-Based Repression of RYBP and its PRC1-Dependent and -Independent Functions. Cell Stem Cell 2017, 20, 462–477.e6. [Google Scholar] [CrossRef] [Green Version]
- Lanzuolo, C.; Orlando, V. Memories from the Polycomb Group Proteins. Annu. Rev. Genet. 2012, 46, 561–589. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yap, K.L.; Zhou, M.-M. Structure and Mechanisms of Lysine Methylation Recognition by the Chromodomain in Gene Transcription. Biochemistry 2011, 50, 1966–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb Group Proteins Ring1A/B Link Ubiquitylation of Histone H2A to Heritable Gene Silencing and X Inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Cao, R.; Tsukada, Y.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef]
- Wilkinson, F.H.; Park, K.; Atchison, M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 2006, 103, 19296–19301. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical Recruitment of Polycomb Group Silencing Complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef]
- Vandamme, J.; Völkel, P.; Rosnoblet, C.; Le Faou, P.; Angrand, P.-O. Interaction Proteomics Analysis of Polycomb Proteins Defines Distinct PRC1 Complexes in Mammalian Cells. Mol. Cell. Proteom. 2011, 10, M110.002642. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, E.; Marcos-Gutiérrez, C.; del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawa, C.; Yoshikawa, T.; Matsuda-Suzuki, F.; Deléhouzée, S.; Goto, M.; Watanabe, H.; Sawada, J.; Kataoka, K.; Handa, H. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 2002, 277, 22484–22490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlisio, S.; Halperin, T.; Vidal, M.; Nevins, J.R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002, 21, 5775–5786. [Google Scholar] [CrossRef]
- Trimarchi, J.M.; Fairchild, B.; Wen, J.; Lees, J.A. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1519–1524. [Google Scholar] [CrossRef]
- Ogawa, H.; Ishiguro, K.-I.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A Complex with Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef]
- Stielow, B.; Finkernagel, F.; Stiewe, T.; Nist, A.; Suske, G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLOS Genet. 2018, 14, e1007193. [Google Scholar] [CrossRef]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Kahn, T.G.; Dorafshan, E.; Schultheis, D.; Zare, A.; Stenberg, P.; Reim, I.; Pirrotta, V.; Schwartz, Y.B. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res. 2016, 44, 10132–10149. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Isono, K.; Kondo, K.; Endo, T.A.; Itohara, S.; Vidal, M.; Koseki, H. Polycomb Potentiates Meis2 Activation in Midbrain by Mediating Interaction of the Promoter with a Tissue-Specific Enhancer. Dev. Cell 2014, 28, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, G. A RING to Rule Them All: RING1 as Silencer and Activator. Dev. Cell 2014, 28, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Huang, Y.; Zhang, J.; Liu, M.; Ji, H.; Wang, C.; Cao, N.; Li, C.; Xia, Y.; Jiang, Q.; et al. Polycomb group RING finger protein 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 2017, jbc.M117.804054. [Google Scholar] [CrossRef] [Green Version]
- Scelfo, A.; Fernández-Pérez, D.; Tamburri, S.; Zanotti, M.; Lavarone, E.; Soldi, M.; Bonaldi, T.; Ferrari, K.J.; Pasini, D. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol. Cell 2019, 74, 1037–1052.e7. [Google Scholar] [CrossRef] [Green Version]
- Bajusz, I.; Kovács, G.; Pirity, M. From Flies to Mice: The Emerging Role of Non-Canonical PRC1 Members in Mammalian Development. Epigenomes 2018, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Junco, S.E.; Wang, R.; Gaipa, J.C.; Taylor, A.B.; Schirf, V.; Gearhart, M.D.; Bardwell, V.J.; Demeler, B.; Hart, P.J.; Kim, C.A. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure 2013, 21, 665–671. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A new ubiquitin-like domain in PRC1 Ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genom. 2008, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Boukhaled, G.M.; Cordeiro, B.; Deblois, G.; Dimitrov, V.; Bailey, S.D.; Holowka, T.; Domi, A.; Guak, H.; Chiu, H.-H.C.; Everts, B.; et al. The Transcriptional Repressor Polycomb Group Factor 6, PCGF6, Negatively Regulates Dendritic Cell Activation and Promotes Quiescence. Cell Rep. 2016, 16, 1829–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endoh, M.; Endo, T.A.; Shinga, J.; Hayashi, K.; Farcas, A.; Ma, K.-W.; Ito, S.; Sharif, J.; Endoh, T.; Onaga, N.; et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 2017, 6. [Google Scholar] [CrossRef]
- Zdzieblo, D.; Li, X.; Lin, Q.; Zenke, M.; Illich, D.J.; Becker, M.; Müller, A.M. Pcgf6, a Polycomb Group Protein, Regulates Mesodermal Lineage Differentiation in Murine ESCs and Functions in iPS Reprogramming. Stem Cells 2014, 32, 3112–3125. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, C.; Li, F.; Jia, L.; Zeng, P.; Li, J.; Tan, J.; Sun, T.; Jiang, S.; Wang, J.; et al. PCGF6 regulates stem cell pluripotency as a transcription activator via super-enhancer dependent chromatin interactions. Protein Cell 2019, 10, 709–725. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Tong, H.; Huang, Y.; Yan, Y.; Teng, H.; Xia, Y.; Jiang, Q.; Qin, J. Essential Role for Polycomb Group Protein Pcgf6 in Embryonic Stem Cell Maintenance and a Noncanonical Polycomb Repressive Complex 1 (PRC1) Integrity. J. Biol. Chem. 2017, 292, 2773–2784. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.-S.; Chang, K.-Y.; Dang, J.; Rana, T.M. Polycomb Group Protein Pcgf6 Acts as a Master Regulator to Maintain Embryonic Stem Cell Identity. Sci. Rep. 2016, 6, 26899. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Wang, J.; He, L.; Lin, Y.; Wu, J. Knockdown of polycomb-group RING finger 6 modulates mouse male germ cell differentiation in vitro. Cell. Physiol. Biochem. 2015, 35, 339–352. [Google Scholar] [CrossRef]
- Pohlers, M.; Truss, M.; Frede, U.; Scholz, A.; Strehle, M.; Kuban, R.-J.; Hoffmann, B.; Morkel, M.; Birchmeier, C.; Hagemeier, C. A Role for E2F6 in the Restriction of Male-Germ-Cell-Specific Gene Expression. Curr. Biol. 2005, 15, 1051–1057. [Google Scholar] [CrossRef] [Green Version]
- Maeda, I.; Okamura, D.; Tokitake, Y.; Ikeda, M.; Kawaguchi, H.; Mise, N.; Abe, K.; Noce, T.; Okuda, A.; Matsui, Y. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 2013, 4, 1754. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, A.; Hirasaki, M.; Hishida, T.; Wu, J.; Okamura, D.; Ueda, A.; Nishimoto, M.; Nakachi, Y.; Mizuno, Y.; Okazaki, Y.; et al. Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat. Commun. 2016, 7, 11056. [Google Scholar] [CrossRef] [PubMed]
- Okuda, A.; Suzuki, A. Unexpected link between MAX and meiotic onset. Cell Cycle 2016, 15, 2235–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Hirasaki, M.; Okuda, A. Does MAX open up a new avenue for meiotic research? Dev. Growth Differ. 2017, 59, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, C.; Liao, J.; Zhao, D.; Huang, H.; Qin, J.; Lee, T.-L.; Chen, D.; Chan, W.-Y.; Xia, Y. L3MBTL2 regulates chromatin remodeling during spermatogenesis. Cell Death Differ. 2019. [Google Scholar] [CrossRef] [PubMed]
- Endoh, M.; Endo, T.A.; Endoh, T.; Isono, K.; Sharif, J.; Ohara, O.; Toyoda, T.; Ito, T.; Eskeland, R.; Bickmore, W.A.; et al. Histone H2A Mono-Ubiquitination Is a Crucial Step to Mediate PRC1-Dependent Repression of Developmental Genes to Maintain ES Cell Identity. PLoS Genet. 2012, 8, e1002774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Mar Lorente, M.; Marcos-Gutiérrez, C.; Pérez, C.; Schoorlemmer, J.; Ramírez, A.; Magin, T.; Vidal, M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 2000, 127, 5093–5100. [Google Scholar]
- Voncken, J.W.; Roelen, B.A.J.; Roefs, M.; de Vries, S.; Verhoeven, E.; Marino, S.; Deschamps, J.; van Lohuizen, M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 2003, 100, 2468–2473. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, T.; Shirley, L.; John, G.B.; Castrillon, D.H. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007, 45, 413–417. [Google Scholar] [CrossRef] [Green Version]
- Morey, L.; Aloia, L.; Cozzuto, L.; Benitah, S.A.; Di Croce, L. RYBP and Cbx7 Define Specific Biological Functions of Polycomb Complexes in Mouse Embryonic Stem Cells. Cell Rep. 2013, 3, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, R.M.; Fagoonee, S.; Papa, A.; Webster, K.; Altruda, F.; Nishinakamura, R.; Chai, L.; Pandolfi, P.P. Functional Antagonism between Sall4 and Plzf Defines Germline Progenitors. Cell Stem Cell 2012, 10, 284–298. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.M.; Finger, J.H.; Kadin, J.A.; Richardson, J.E.; Ringwald, M. The gene expression database for mouse development (GXD): Putting developmental expression information at your fingertips. Dev. Dyn. 2014, 243, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Gassei, K.; Orwig, K.E. SALL4 Expression in Gonocytes and Spermatogonial Clones of Postnatal Mouse Testes. PLoS ONE 2013, 8, e53976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Liu, M.; Ji, H.; Zhu, Y.; Wang, C.; Huang, Y.; Ma, X.; Xing, G.; Xia, Y.; Jiang, Q.; et al. The polycomb group protein Yaf2 regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. J. Biol. Chem. 2018, 293, 12793–12804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujhelly, O.; Szabo, V.; Kovacs, G.; Vajda, F.; Mallok, S.; Prorok, J.; Acsai, K.; Hegedus, Z.; Krebs, S.; Dinnyes, A.; et al. Lack of Rybp in Mouse Embryonic Stem Cells Impairs Cardiac Differentiation. Stem Cells Dev. 2015, 24, 2193–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Stoop, P.; Boutsma, E.A.; Hulsman, D.; Noback, S.; Heimerikx, M.; Kerkhoven, R.M.; Voncken, J.W.; Wessels, L.F.A.; van Lohuizen, M. Ubiquitin E3 Ligase Ring1b/Rnf2 of Polycomb Repressive Complex 1 Contributes to Stable Maintenance of Mouse Embryonic Stem Cells. PLoS ONE 2008, 3, e2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hod-Dvorai, R.; Jacob, E.; Boyko, Y.; Avni, O. The binding activity of Mel-18 at the Il17a promoter is regulated by the integrated signals of the TCR and polarizing cytokines. Eur. J. Immunol. 2011, 41, 2424–2435. [Google Scholar] [CrossRef]
- Morey, L.; Santanach, A.; Blanco, E.; Aloia, L.; Nora, E.P.; Bruneau, B.G.; Di Croce, L. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell 2015, 17, 300–315. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Tseng, W.-C.; Fan, X.; Ball, R.; Dougan, S.T. Extraembryonic Signals under the Control of MGA, Max, and Smad4 Are Required for Dorsoventral Patterning. Dev. Cell 2014, 28, 322–334. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Chen, J.; Zhang, Y.; Munisha, M.; Dougan, S.; Sun, Y. Mga Modulates Bmpr1a Activity by Antagonizing Bs69 in Zebrafish. Front. Cell Dev. Biol. 2018, 6, 126. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, Y.; Feng, X.; Liao, S.; Wang, X.; Gan, H.; Wang, L.; Lin, X.; Han, C. BMP4 Cooperates with Retinoic Acid to Induce the Expression of Differentiation Markers in Cultured Mouse Spermatogonia. Stem Cells Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Baleato, R.M.; Aitken, R.J.; Roman, S.D. Vitamin A regulation of BMP4 expression in the male germ line. Dev. Biol. 2005, 286, 78–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyauchi, H.; Ohta, H.; Nagaoka, S.; Nakaki, F.; Sasaki, K.; Hayashi, K.; Yabuta, Y.; Nakamura, T.; Yamamoto, T.; Saitou, M. Bone morphogenetic protein and retinoic acid synergistically specify female germ-cell fate in mice. EMBO J. 2017, 36, 3100–3119. [Google Scholar] [CrossRef] [PubMed]
- Kalenik, J.L.; Chen, D.; Bradley, M.E.; Chen, S.J.; Lee, T.C. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997, 25, 843–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eid, A.; Torres-Padilla, M.-E. Characterization of non-canonical Polycomb Repressive Complex 1 subunits during early mouse embryogenesis. Epigenetics 2016, 11, 389–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirity, M.K.; Locker, J.; Schreiber-Agus, N. Rybp/DEDAF Is Required for Early Postimplantation and for Central Nervous System Development. Mol. Cell. Biol. 2005, 25, 7193–7202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, K.A.; Hage, W.J. Clonal Analysis of the Origin of Primordial Germ Cells in the Mouse. Ciba Found. Symp. 2007, 182, 68–91. [Google Scholar]
- Hayashi, K.; de Sousa Lopes, S.M.C.; Surani, M.A. Germ Cell Specification in Mice. Science 2007, 316, 394–396. [Google Scholar] [CrossRef]
- Hisada, K.; Sánchez, C.; Endo, T.A.; Endoh, M.; Román-Trufero, M.; Sharif, J.; Koseki, H.; Vidal, M. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 2012, 32, 1139–1149. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, G.; Szabo, V.; Pirity, M.K. Absence of Rybp Compromises Neural Differentiation of Embryonic Stem Cells. Stem Cells Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hermann, B.P.; Mutoji, K.N.; Velte, E.K.; Ko, D.; Oatley, J.M.; Geyer, C.B.; McCarrey, J.R. Transcriptional and Translational Heterogeneity among Neonatal Mouse Spermatogonia1. Biol. Reprod. 2015, 92, 54. [Google Scholar] [CrossRef]
- Buehr, M.; Nichols, J.; Stenhouse, F.; Mountford, P.; Greenhalgh, C.J.; Kantachuvesiri, S.; Brooker, G.; Mullins, J.; Smith, A.G. Rapid Loss of Oct-4 and Pluripotency in Cultured Rodent Blastocysts and Derivative Cell Lines1. Biol. Reprod. 2003, 68, 222–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwaka, T.P.; Thomson, J.A. A germ cell origin of embryonic stem cells? Development 2005, 132, 227–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, L.-F.; Surani, M.A.; Jaenisch, R.; Zwaka, T.P. Blimp1 Expression Predicts Embryonic Stem Cell Development In Vitro. Curr. Biol. 2011, 21, 1759–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurimoto, K.; Yabuta, Y.; Ohinata, Y.; Shigeta, M.; Yamanaka, K.; Saitou, M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008, 22, 1617–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubner, K.; Fuhrmann, G.; Christenson, L.K.; Kehler, J.; Reinbold, R.; De La Fuente, R.; Wood, J.; Strauss, J.F.; Boiani, M.; Schöler, H.R. Derivation of Oocytes from Mouse Embryonic Stem Cells. Science 2003, 300, 1251–1256. [Google Scholar] [CrossRef] [Green Version]
- Geijsen, N.; Horoschak, M.; Kim, K.; Gribnau, J.; Eggan, K.; Daley, G.Q. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004, 427, 148–154. [Google Scholar] [CrossRef]
- Clark, A.T.; Bodnar, M.S.; Fox, M.; Rodriquez, R.T.; Abeyta, M.J.; Firpo, M.T.; Pera, R.A.R. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004, 13, 727–739. [Google Scholar] [CrossRef] [Green Version]
- Donovan, P.J.; de Miguel, M.P. Turning germ cells into stem cells. Curr. Opin. Genet. Dev. 2003, 13, 463–471. [Google Scholar] [CrossRef]
- Carstea, A.C.; Pirity, M.K.; Dinnyes, A. Germline competence of mouse ES and iPS cell lines: Chimera technologies and genetic background. World J. Stem Cells 2009, 1, 22. [Google Scholar] [CrossRef]
- Keskintepe, L.; Norris, K.; Pacholczyk, G.; Dederscheck, S.M.; Eroglu, A. Derivation and comparison of C57BL/6 embryonic stem cells to a widely used 129 embryonic stem cell line. Transgenic Res. 2007, 16, 751–758. [Google Scholar] [CrossRef]
- Kleinsmith, L.J.; Pierce, G.B. Multipotentiality Of Single Embryonal Carcinoma Cells. Cancer Res. 1964, 24, 1544–1551. [Google Scholar] [PubMed]
- Dixon, F.S.; Moore, R.A. Tumors of the Male Sex Organs. Ann. Intern. Med. 1954, 40, 828. [Google Scholar] [CrossRef]
- Stevens, L.C.; Little, C.C. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proc. Natl. Acad. Sci. USA 1954, 40, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahan, B.W.; Ephrussi, B. Developmental potentialities of clonal in vitro cultures of mouse testicular teratoma. J. Natl. Cancer Inst. 1970, 44, 1015–1036. [Google Scholar]
- Stevens, L.C. Experimental production of testicular teratomas in mice of strains 129, A/He, and their F1 hybrids. J. Natl. Cancer Inst. 1970, 44, 923–929. [Google Scholar]
- Smith, A.G. Embryo-Derived Stem Cells: Of Mice and Men. Annu. Rev. Cell Dev. Biol. 2001, 17, 435–462. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Sekinaka, T.; Tando, Y.; Okamura, D.; Tanaka, K.; Ito-Matsuoka, Y.; Takehara, A.; Yaegashi, N.; Matsui, Y. Derivation of pluripotent stem cells from nascent undifferentiated teratoma. Dev. Biol. 2019, 446, 43–55. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [Green Version]
- Liu, A.; Cheng, L.; Du, J.; Peng, Y.; Allan, R.W.; Wei, L.; Li, J.; Cao, D. Diagnostic Utility of Novel Stem Cell Markers SALL4, OCT4, NANOG, SOX2, UTF1, and TCL1 in Primary Mediastinal Germ Cell Tumors. Am. J. Surg. Pathol. 2010, 34, 1. [Google Scholar] [CrossRef]
- Cao, D.; Guo, S.; Allan, R.W.; Molberg, K.H.; Peng, Y. SALL4 Is a Novel Sensitive and Specific Marker of Ovarian Primitive Germ Cell Tumors and Is Particularly Useful in Distinguishing Yolk Sac Tumor From Clear Cell Carcinoma. Am. J. Surg. Pathol. 2009, 33, 894–904. [Google Scholar] [CrossRef]
- Miettinen, M.; Wang, Z.; McCue, P.A.; Sarlomo-Rikala, M.; Rys, J.; Biernat, W.; Lasota, J.; Lee, Y.-S. SALL4 Expression in Germ Cell and Non–Germ Cell Tumors. Am. J. Surg. Pathol. 2014, 38, 410–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFarlane, R.J.; Wakeman, J.A. Meiosis-like Functions in Oncogenesis: A New View of Cancer. Cancer Res. 2017, 77, 5712–5716. [Google Scholar] [CrossRef] [PubMed]
- Huberts, D.H.E.W.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 520–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neira, J.L.; Román-Trufero, M.; Contreras, L.M.; Prieto, J.; Singh, G.; Barrera, F.N.; Renart, M.L.; Vidal, M. The Transcriptional Repressor RYBP Is a Natively Unfolded Protein Which Folds upon Binding to DNA. Biochemistry 2009, 48, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Schickling, O.; Peter, M.E.; Lenardo, M.J. The Death Effector Domain-associated Factor Plays Distinct Regulatory Roles in the Nucleus and Cytoplasm. J. Biol. Chem. 2001, 276, 31945–31952. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Zhang, X.; Li, M.; Ma, X.; Huang, B.; Chen, H.; Chen, D. Proapoptotic RYBP interacts with FANK1 and induces tumor cell apoptosis through the AP-1 signaling pathway. Cell. Signal. 2016, 28, 779–787. [Google Scholar] [CrossRef]
- Wang, H.; Song, W.; Hu, T.; Zhang, N.; Miao, S.; Zong, S.; Wang, L. Fank1 interacts with Jab1 and regulates cell apoptosis via the AP-1 pathway. Cell. Mol. Life Sci. 2011, 68, 2129–2139. [Google Scholar] [CrossRef]
- Zheng, Z.; Zheng, H.; Yan, W. Fank1 is a testis-specific gene encoding a nuclear protein exclusively expressed during the transition from the meiotic to the haploid phase of spermatogenesis. Gene Expr. Patterns 2007, 7, 777–783. [Google Scholar] [CrossRef]
- Hwang, K.-C.; Park, S.-Y.; Park, S.-P.; Lim, J.H.; Cui, X.-S.; Kim, N.-H. Specific maternal transcripts in bovine oocytes and cleavaged embryos: Identification with novel DDRT-PCR methods. Mol. Reprod. Dev. 2005, 71, 275–283. [Google Scholar] [CrossRef]
- Dong, W.-W.; Huang, H.-L.; Yang, W.; Liu, J.; Yu, Y.; Zhou, S.-L.; Wang, W.; Lv, X.-C.; Li, Z.-Y.; Zhang, M.-Y.; et al. Testis-specific Fank1 gene in knockdown mice produces oligospermia via apoptosis. Asian J. Androl. 2014, 16, 124. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Joza, N.; Tasdemir, E.; Maiuri, M.C.; Hengartner, M.; Abrams, J.M.; Tavernarakis, N.; Penninger, J.; Madeo, F.; Kroemer, G. No death without life: Vital functions of apoptotic effectors. Cell Death Differ. 2008, 15, 1113–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fereres, S.; Simón, R.; Mohd-Sarip, A.; Verrijzer, C.P.; Busturia, A. dRYBP Counteracts Chromatin-Dependent Activation and Repression of Transcription. PLoS ONE 2014, 9, e113255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calés, C.; Pavón, L.; Starowicz, K.; Pérez, C.; Bravo, M.; Ikawa, T.; Koseki, H.; Vidal, M. Role of Polycomb RYBP in Maintaining the B-1-to-B-2 B-Cell Lineage Switch in Adult Hematopoiesis. Mol. Cell. Biol. 2016, 36, 900–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparicio, R.; Neyen, C.; Lemaitre, B.; Busturia, A. dRYBP Contributes to the Negative Regulation of the Drosophila Imd Pathway. PLoS ONE 2013, 8, e62052. [Google Scholar] [CrossRef] [Green Version]
- Satijn, D.P.; Gunster, M.J.; van der Vlag, J.; Hamer, K.M.; Schul, W.; Alkema, M.J.; Saurin, A.J.; Freemont, P.S.; van Driel, R.; Otte, A.P. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 1997, 17, 4105–4113. [Google Scholar] [CrossRef] [Green Version]
- Atchison, L.; Ghias, A.; Wilkinson, F.; Bonini, N.; Atchison, M.L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003, 22, 1347–1358. [Google Scholar] [CrossRef]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin Compaction by a Polycomb Group Protein Complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef] [Green Version]
- Lund, A.H.; van Lohuizen, M. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 2004, 16, 239–246. [Google Scholar] [CrossRef]
- Taherbhoy, A.M.; Huang, O.W.; Cochran, A.G. BMI1–RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 2015, 6, 7621. [Google Scholar] [CrossRef]
- Czypionka, A.; Ruiz de los Paños, O.; Mateu, M.G.; Barrera, F.N.; Hurtado-Gómez, E.; Gómez, J.; Vidal, M.; Neira, J.L. The Isolated C-Terminal Domain of Ring1B Is a Dimer Made of Stable, Well-Structured Monomers. Biochemistry 2007, 46, 12764–12776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezsonova, I.; Walker, J.R.; Bacik, J.P.; Duan, S.; Dhe-Paganon, S.; Arrowsmith, C.H. Ring1B Contains a Ubiquitin-Like Docking Module for Interaction with Cbx Proteins. Biochemistry 2009, 48, 10542–10548. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Wilkinson, F.H.; Colavita, K.; Fennelly, C.; Atchison, M.L. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014, 42, 2208–2223. [Google Scholar] [CrossRef] [PubMed]
- Kahn, T.G.; Stenberg, P.; Pirrotta, V.; Schwartz, Y.B. Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements. PLoS Genet. 2014, 10, e1004495. [Google Scholar] [CrossRef] [Green Version]
- Donohoe, M.E.; Zhang, X.; McGinnis, L.; Biggers, J.; Li, E.; Shi, Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999, 19, 7237–7244. [Google Scholar] [CrossRef] [Green Version]
- Mihaly, J.; Mishra, R.K.; Karch, F. A conserved sequence motif in Polycomb-response elements. Mol. Cell 1998, 1, 1065–1066. [Google Scholar] [CrossRef]
- Simon, J.; Chiang, A.; Bender, W.; Shimell, M.J.; O’Connor, M. Elements of the Drosophila Bithorax Complex That Mediate Repression by Polycomb Group Products. Dev. Biol. 1993, 158, 131–144. [Google Scholar] [CrossRef]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A Region of the Human HOXD Cluster that Confers Polycomb-Group Responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [Green Version]
- Sing, A.; Pannell, D.; Karaiskakis, A.; Sturgeon, K.; Djabali, M.; Ellis, J.; Lipshitz, H.D.; Cordes, S.P. A Vertebrate Polycomb Response Element Governs Segmentation of the Posterior Hindbrain. Cell 2009, 138, 885–897. [Google Scholar] [CrossRef] [Green Version]
- Mohd-Sarip, A.; Cléard, F.; Mishra, R.K.; Karch, F.; Verrijzer, C.P. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005, 19, 1755–1760. [Google Scholar] [CrossRef] [Green Version]
- Klymenko, T.; Papp, B.; Fischle, W.; Köcher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Müller, J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsch, C.; Brown, J.L.; Kassis, J.A.; Müller, J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 1999, 126, 3905–3913. [Google Scholar] [PubMed]
- Shimell, M.J.; Peterson, A.J.; Burr, J.; Simon, J.A.; O’Connor, M.B. Functional Analysis of Repressor Binding Sites in the iab-2 Regulatory Region of the abdominal—A Homeotic Gene. Dev. Biol. 2000, 218, 38–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busturia, A.; Lloyd, A.; Bejarano, F.; Zavortink, M.; Xin, H.; Sakonju, S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 2001, 128, 2163–2173. [Google Scholar]
- Mishra, R.K.; Mihaly, J.; Barges, S.; Spierer, A.; Karch, F.; Hagstrom, K.; Schweinsberg, S.E.; Schedl, P. The iab-7 Polycomb Response Element Maps to a Nucleosome-Free Region of Chromatin and Requires Both GAGA and Pleiohomeotic for Silencing Activity. Mol. Cell. Biol. 2001, 21, 1311–1318. [Google Scholar] [CrossRef] [Green Version]
- Kozma, G.; Bender, W.; Sipos, L. Replacement of a Drosophila Polycomb response element core, and in situ analysis of its DNA motifs. Mol. Genet. Genom. 2008, 279, 595–603. [Google Scholar] [CrossRef]
- Chagraoui, H.; Kristiansen, M.S.; Ruiz, J.P.; Serra-Barros, A.; Richter, J.; Hall-Ponselé, E.; Gray, N.; Waithe, D.; Clark, K.; Hublitz, P.; et al. SCL/TAL1 cooperates with Polycomb RYBP-PRC1 to suppress alternative lineages in blood-fated cells. Nat. Commun. 2018, 9, 5375. [Google Scholar] [CrossRef]
- Cohen, I.; Zhao, D.; Bar, C.; Valdes, V.J.; Dauber-Decker, K.L.; Nguyen, M.B.; Nakayama, M.; Rendl, M.; Bickmore, W.A.; Koseki, H.; et al. PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell 2018, 22, 726–739.e7. [Google Scholar] [CrossRef] [Green Version]
- Velasco, G.; Hube, F.; Rollin, J.; Neuillet, D.; Philippe, C.; Bouzinba-Segard, H.; Galvani, A.; Viegas-Pequignot, E.; Francastel, C. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc. Natl. Acad. Sci. USA 2010, 107, 9281–9286. [Google Scholar] [CrossRef] [Green Version]
- Storre, J.; Elsässer, H.-P.; Fuchs, M.; Ullmann, D.; Livingston, D.M.; Gaubatz, S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002, 3, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Arrigoni, R.; Alam, S.L.; Wamstad, J.A.; Bardwell, V.J.; Sundquist, W.I.; Schreiber-Agus, N. The Polycomb-associated protein Rybp is A ubiquitin binding protein. FEBS Lett. 2006, 580, 6233–6241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchwald, G.; van der Stoop, P.; Weichenrieder, O.; Perrakis, A.; van Lohuizen, M.; Sixma, T.K. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006, 25, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Bentley, M.L.; Corn, J.E.; Dong, K.C.; Phung, Q.; Cheung, T.K.; Cochran, A.G. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011, 30, 3285–3297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Alam, S.L.; Meyer, H.H.; Payne, M.; Stemmler, T.L.; Davis, D.R.; Sundquist, W.I. Structure and Ubiquitin Interactions of the Conserved Zinc Finger Domain of Npl4. J. Biol. Chem. 2003, 278, 20225–20234. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhang, S.; Zhao, W.; Hou, C.; Ma, X.; Li, X.; Huang, B.; Chen, H.; Chen, D. RYBP modulates stability and function of Ring1B through targeting UBE3A. FASEB J. 2019, 33, 683–695. [Google Scholar] [CrossRef]
- Reik, W.; Surani, M.A. Germline and Pluripotent Stem Cells. Cold Spring Harb. Perspect. Biol. 2015, 7, a019422. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.A.M.; Strickfaden, H.; Lee, B.L.; Spyracopoulos, L.; Hendzel, M.J. RYBP Is a K63-Ubiquitin-Chain-Binding Protein that Inhibits Homologous Recombination Repair. Cell Rep. 2018, 22, 383–395. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, D.L.C.; Snoek, T.; Mullin, N.P.; Yates, A.; Bezstarosti, K.; Demmers, J.; Chambers, I.; Poot, R.A. An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 2010, 6, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Bowles, J.; Koopman, P. Retinoic acid, meiosis and germ cell fate in mammals. Development 2007, 134, 3401–3411. [Google Scholar] [CrossRef] [Green Version]
- Toyooka, Y.; Tsunekawa, N.; Akasu, R.; Noce, T. Embryonic stem cells can form germ cells in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 11457–11462. [Google Scholar] [CrossRef] [Green Version]
- West, J.A.; Park, I.-H.; Daley, G.Q.; Geijsen, N. In vitro generation of germ cells from murine embryonic stem cells. Nat. Protoc. 2006, 1, 2026–2036. [Google Scholar] [CrossRef] [PubMed]
- Toyooka, Y.; Tsunekawa, N.; Takahashi, Y.; Matsui, Y.; Satoh, M.; Noce, T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000, 93, 139–149. [Google Scholar] [CrossRef]
- BAUGHMAN, J.M.; Geijsen, N. In Vitro Generation of Germ Cells: New Techniques to Solve Current Issues. Ann. N.Y. Acad. Sci. 2005, 1061, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Kee, K.; Gonsalves, J.M.; Clark, A.T.; Pera, R.A.R. Bone Morphogenetic Proteins Induce Germ Cell Differentiation from Human Embryonic Stem Cells. Stem Cells Dev. 2006, 15, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Nayernia, K.; Li, M.; Jaroszynski, L.; Khusainov, R.; Wulf, G.; Schwandt, I.; Korabiowska, M.; Michelmann, H.W.; Meinhardt, A.; Engel, W. Stem cell based therapeutical approach of male infertility by teratocarcinoma derived germ cells. Hum. Mol. Genet. 2004, 13, 1451–1460. [Google Scholar] [CrossRef]
- Nayernia, K.; Lee, J.H.; Drusenheimer, N.; Nolte, J.; Wulf, G.; Dressel, R.; Gromoll, J.; Engel, W. Derivation of male germ cells from bone marrow stem cells. Lab. Investig. 2006, 86, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Nayernia, K.; Nolte, J.; Michelmann, H.W.; Lee, J.H.; Rathsack, K.; Drusenheimer, N.; Dev, A.; Wulf, G.; Ehrmann, I.E.; Elliott, D.J.; et al. In Vitro-Differentiated Embryonic Stem Cells Give Rise to Male Gametes that Can Generate Offspring Mice. Dev. Cell 2006, 11, 125–132. [Google Scholar] [CrossRef] [Green Version]
- De Kumar, B.; Parrish, M.E.; Slaughter, B.D.; Unruh, J.R.; Gogol, M.; Seidel, C.; Paulson, A.; Li, H.; Gaudenz, K.; Peak, A.; et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genom. Res. 2015, 25, 1229–1243. [Google Scholar] [CrossRef] [Green Version]
- Griswold, M.D.; Hogarth, C.A.; Bowles, J.; Koopman, P. Initiating Meiosis: The Case for Retinoic Acid1. Biol. Reprod. 2012, 86, 35. [Google Scholar] [CrossRef]
- Anderson, E.L.; Baltus, A.E.; Roepers-Gajadien, H.L.; Hassold, T.J.; de Rooij, D.G.; van Pelt, A.M.M.; Page, D.C. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 14976–14980. [Google Scholar] [CrossRef] [Green Version]
- Sucov, H.M.; Murakami, K.K.; Evans, R.M. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc. Natl. Acad. Sci. USA 1990, 87, 5392–5396. [Google Scholar] [CrossRef] [Green Version]
- Sharov, A.A.; Masui, S.; Sharova, L.V.; Piao, Y.; Aiba, K.; Matoba, R.; Xin, L.; Niwa, H.; Ko, M.S. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genom. 2008, 9, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog Is the Gateway to the Pluripotent Ground State. Cell 2009, 138, 722–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, A.T.; Rodriguez, R.T.; Bodnar, M.S.; Abeyta, M.J.; Cedars, M.I.; Turek, P.J.; Firpo, M.T.; Reijo Pera, R.A. Human STELLAR, NANOG, and GDF3 Genes Are Expressed in Pluripotent Cells and Map to Chromosome 12p13, a Hotspot for Teratocarcinoma. Stem Cells 2004, 22, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Kuijk, E.W.; de Gier, J.; Chuva de Sousa Lopes, S.M.; Chambers, I.; van Pelt, A.M.M.; Colenbrander, B.; Roelen, B.A.J. A Distinct Expression Pattern in Mammalian Testes Indicates a Conserved Role for NANOG in Spermatogenesis. PLoS ONE 2010, 5, e10987. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahony, S.; Mazzoni, E.O.; McCuine, S.; Young, R.A.; Wichterle, H.; Gifford, D.K. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genom. Biol. 2011, 12, R2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santanach, A.; Blanco, E.; Jiang, H.; Molloy, K.R.; Sansó, M.; LaCava, J.; Morey, L.; Di Croce, L. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat. Commun. 2017, 8, 1235. [Google Scholar] [CrossRef]
- Kogo, H.; Tsutsumi, M.; Inagaki, H.; Ohye, T.; Kiyonari, H.; Kurahashi, H. HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells 2012, 17, 897–912. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Tilly, J.L. Epigenetic status determines germ cell meiotic commitment in embryonic and postnatal mammalian gonads. Cell Cycle 2010, 9, 339–349. [Google Scholar] [CrossRef]
- Garchon, H.J. The Xlr (X-linked lymphocyte regulated) gene family (a candidate locus for an X-linked primary immune deficiency). Immunodefic. Rev. 1991, 2, 283–302. [Google Scholar] [PubMed]
- Escalier, D.; Eloy, L.; Garchon, H.-J. Sex-Specific Gene Expression During Meiotic Prophase I: Xlr (X Linked, Lymphocyte Regulated), Not Its Male Homologue Xmr (Xlr Related, Meiosis Regulated), Is Expressed in Mouse Oocytes. Biol. Reprod. 2002, 67, 1646–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Qu, P.; Zhou, C.; Liu, X.; Ma, X.; Wang, M.; Wang, Y.; Su, J.; Liu, J.; Zhang, Y. MicroRNA-125b is a key epigenetic regulatory factor that promotes nuclear transfer reprogramming. J. Biol. Chem. 2017, 292, 15916–15926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirity, M.K.; Wang, W.-L.; Wolf, L.V.; Tamm, E.R.; Schreiber-Agus, N.; Cvekl, A. Rybp, a polycomb complex-associated protein, is required for mouse eye development. BMC Dev. Biol. 2007, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene Name | Transcriptome (Rybp+/+/Rybp-/-) Raw Counts | Fold Change | RYBP ChIP-seq (GSM1041375) | RING1B ChIP-seq (GSM1041372) | PCGF6 ChIP-seq (GSE84905) | RAR ChIP-seq (GSM482749) |
|---|---|---|---|---|---|---|
| Adh1 | 0/0.7 | Inf | - | - | - | - |
| Adh4 | 0/5.65 | Inf | - | - | - | + |
| Rarb | 0/2.12 | Inf | - | + | - | + |
| Rxrg | 0/1.41 | Inf | - | + | + | + |
| Crabp1 | 36.8/218.5 | 5.94 | + | + | - | - |
| Rdh14 | 4.2/13.4 | 3.166 | - | - | - | - |
| Rxra | 29.7/38.2 | 1.285 | - | + | - | + |
| Gene Name | Transcriptome (Rybp+/+/Rybp-/-) Raw Counts | Fold Change | RYBP ChIP-seq (GSM1041375) | RING1B ChIP-seq (GSM1041372) | PCGF6 ChIP-seq (GSE84905) | RAR ChIP-seq (GSM482749) |
|---|---|---|---|---|---|---|
| Xlr4a | 0/42.42 | Inf | - | - | - | - |
| Xlr4b | 0/42.43 | Inf | - | - | - | - |
| Tex11 | 5.65/223.44 | 39.50 | + | + | + | - |
| Dazl | 31.11/437.69 | 14.068 | + | + | + | - |
| Tex15 | 31.11/280.01 | 9.00 | + | + | - | + |
| Xlr3a | 7.07/63.63 | 9.00 | - | - | - | - |
| Piwil2 | 22.62/201.52 | 8.90 | + | + | + | + |
| Xlr3c | 1.41/12.02 | 8.50 | - | - | - | - |
| Tdrkh | 29.69/250.31 | 8.42 | + | + | + | - |
| Hormad2 | 16.97/110.30 | 6.50 | + | - | - | + |
| Sycp2 | 21.21/137.17 | 6.46 | + | + | + | + |
| Mov10l1 | 55.15/335.87 | 6.090 | + | + | + | + |
| Piwil4 | 5.65/33.94 | 6.00 | - | - | - | + |
| Mael | 87.68/415.77 | 4.74 | + | + | + | + |
| Ddx4 | 151.32/623.66 | 4.120 | + | + | + | + |
| Tdrd1 | 18.38/65.76 | 3.57 | + | + | + | + |
| Smc1b | 100.40/337.28 | 3.35 | + | + | + | + |
| Xlr3b | 9.89/32.52 | 3.285 | - | - | - | - |
| Cpeb1 | 11.31/36.06 | 3.187 | + | + | + | + |
| Stra8 | 48.08/152.73 | 3.176 | - | - | - | + |
| Boll | 7.07/20.5 | 2.90 | + | + | + | + |
| Meioc | 49.49/142.12 | 2.87 | + | + | + | + |
| Asz1 | 2.82/7.77 | 2.75 | + | + | - | + |
| Sycp1 | 251.73/412.95 | 1.640 | + | + | + | - |
| Prdm14 | 52.34/84.14 | 1.60 | - | + | - | + |
| Sycp3 | 322.44/381.83 | 1.184 | + | + | - | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajusz, I.; Henry, S.; Sutus, E.; Kovács, G.; Pirity, M.K. Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription. Genes 2019, 10, 941. https://doi.org/10.3390/genes10110941
Bajusz I, Henry S, Sutus E, Kovács G, Pirity MK. Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription. Genes. 2019; 10(11):941. https://doi.org/10.3390/genes10110941
Chicago/Turabian StyleBajusz, Izabella, Surya Henry, Enikő Sutus, Gergő Kovács, and Melinda K. Pirity. 2019. "Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription" Genes 10, no. 11: 941. https://doi.org/10.3390/genes10110941
APA StyleBajusz, I., Henry, S., Sutus, E., Kovács, G., & Pirity, M. K. (2019). Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription. Genes, 10(11), 941. https://doi.org/10.3390/genes10110941




