Plant PHR Transcription Factors: Put on A Map
Abstract
:1. Introduction
2. PHRs Redundancy and Dimerization
2.1. Cooperation between PHR Family Members
2.2. PHRs Work Together in a Link
3. The Multifunctional Role of PHR1
3.1. PHR1 Affects Plant Immune System
3.2. Metal-Phosphate Relationship Modulated by PHR1
3.3. Double-Faced Role of PHR1 in the Regulation of Sulfate Homeostasis
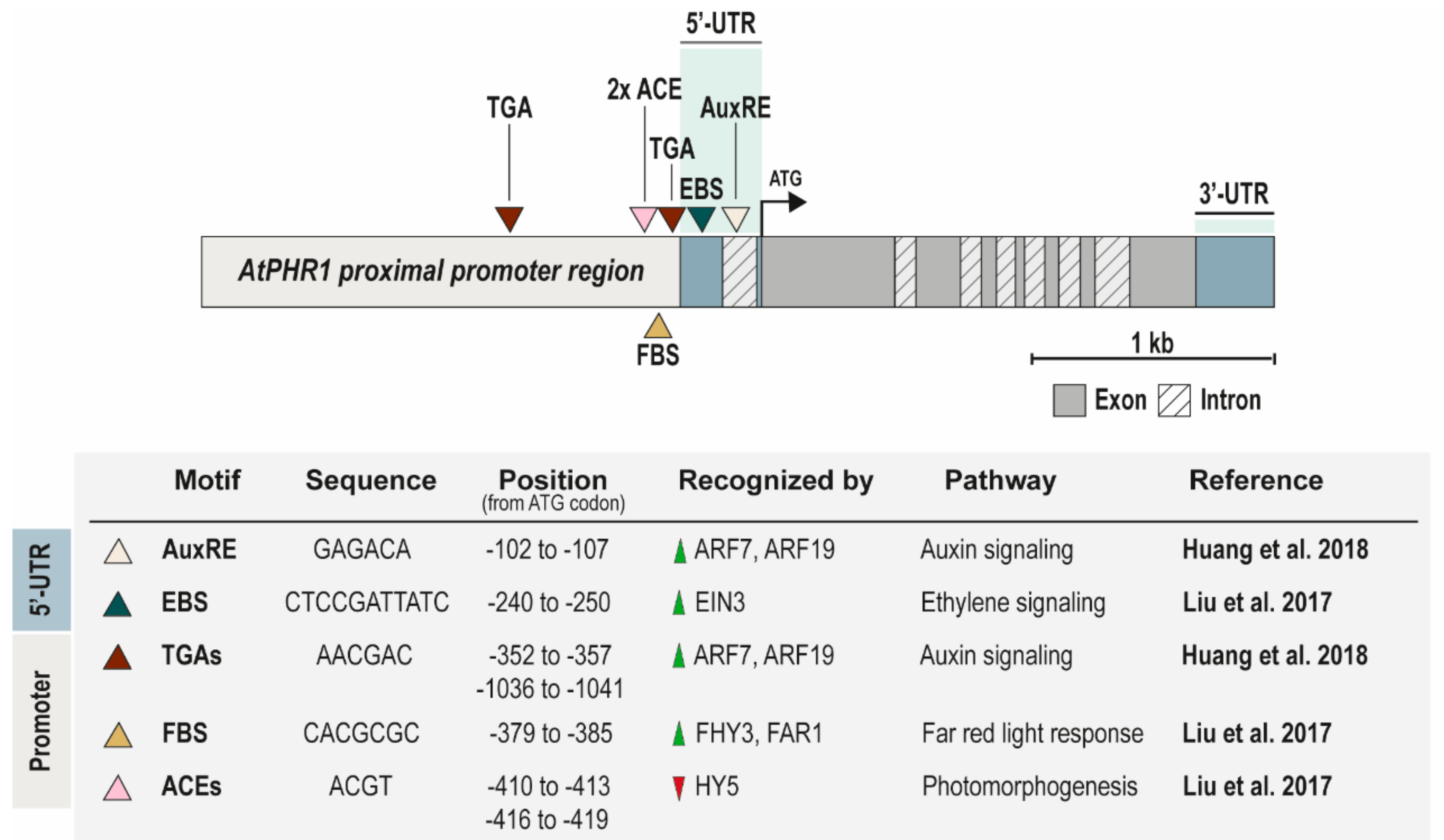
4. Transcriptional Regulation of PHR1 Gene Expression
PHR1 Promoter as a Station for Many Plant TFs
5. PHR1 Post-Translational Modifications
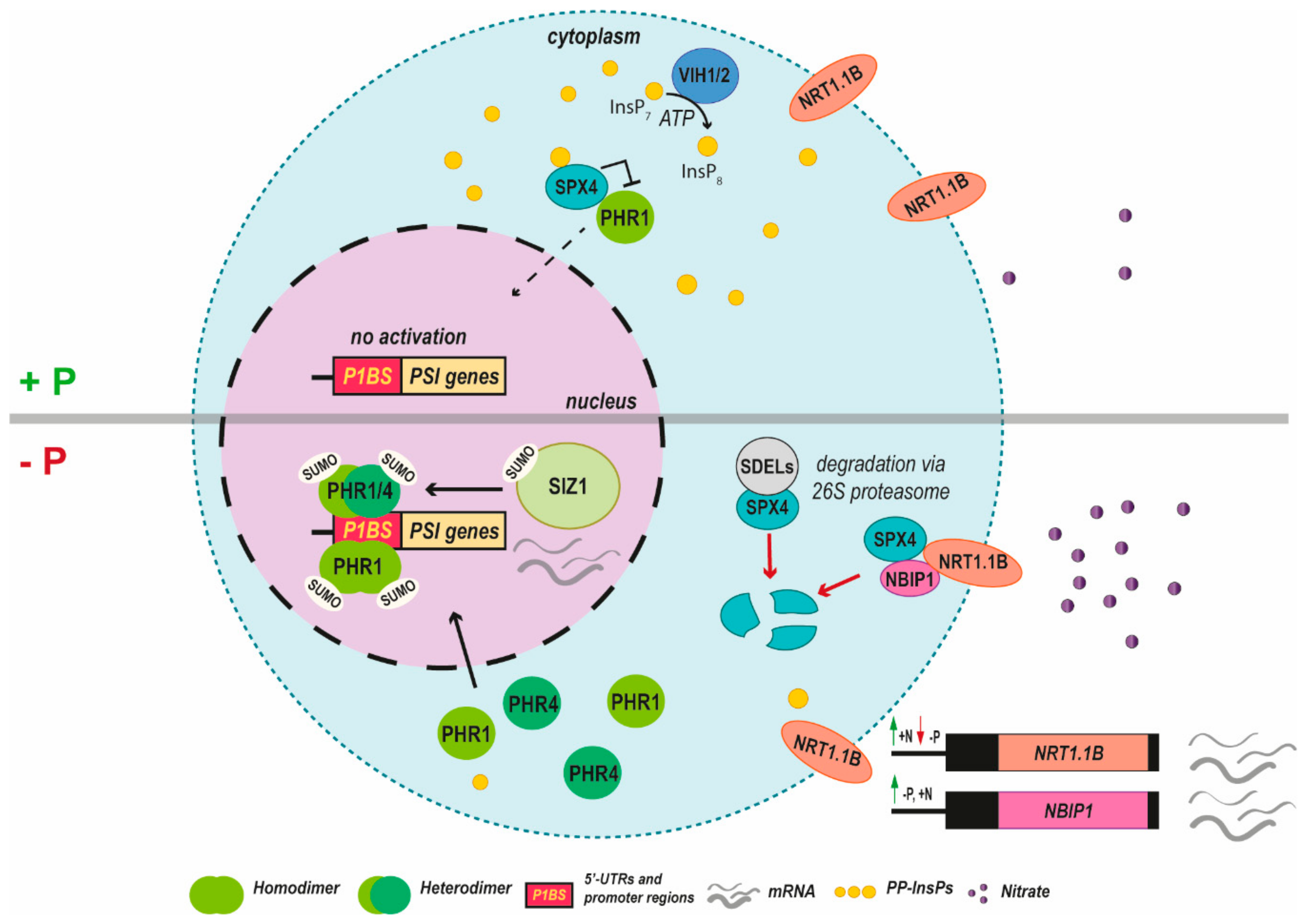
6. PHR1 Meets Nitrogen and Phosphate Sensors
6.1. SPX Proteins Navigate PHR1 in Plant Cells
6.2. Inositol Pyrophosphates (PP-InsPs) as Messenger
6.3. SPX Proteins from the Nitrogen Perspective
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [Green Version]
- Heffer, P.; Prud’homme, M. Nutrients as limited resources: Global trends in fertilizer production and use. In Improving Water and Nutrient-Use Efficiency in Food Production Systems; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 57–78. ISBN 9780813819891. [Google Scholar]
- Shimogawara, K.; Wykoff, D.D.; Usuda, H.; Grossman, A.R. Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. 1999, 120, 685–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wykoff, D.D.; Grossman, A.R.; Weeks, D.P.; Usuda, H.; Shimogawara, K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 1999, 96, 15336–15341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajhaiya, A.K.; Dean, A.P.; Zeef, L.A.; Webster, R.E.; Pittman, J.K. PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii. Plant Physiol. 2016, 170, 1216–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-ares, J. A conserved MYB transcription factor involved in Phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 is involved in Phosphate-starvation signaling and excessive Phosphate accumulation in shoots of plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Wang, Z.; Ren, H.; Shen, C.; Li, Y.; Ling, H.Q.; Wu, C.; Lian, X.; Wu, P. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and Phosphate homeostasis in shoots of rice. Plant J. 2010, 62, 508–517. [Google Scholar] [CrossRef]
- Nilsson, L.; Müller, R.; Nielsen, T.H. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced Phosphate uptake in Arabidopsis thaliana. Plant. Cell Environ. 2007, 30, 1499–1512. [Google Scholar] [CrossRef]
- Pant, B.D.; Burgos, A.; Pant, P.; Cuadros-Inostroza, A.; Willmitzer, L.; Scheible, W.R. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 2015, 66, 1907–1918. [Google Scholar] [CrossRef] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to Phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.S.; Lai, F.J. Functional redundancy of transcription factors explains why most binding targets of a transcription factor are not affected when the transcription factor is knocked out. BMC Syst. Biol. 2015, 9, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuntz, S.G.; Williams, B.A.; Sternberg, P.W.; Wold, B.J. Transcription factor redundancy and tissue-specific regulation: Evidence from functional and physical network connectivity. Genome Res. 2012, 22, 1907–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.; Sun, L.; Isupov, M.N.; Littlechild, J.A.; Wu, X.; Wang, Q.; Wang, Q.; Yang, W.; Wu, Y. Structural basis for the target DNA recognition and binding by the MYB domain of Phosphate starvation response 1. FEBS J. 2019, 286, 2809–2821. [Google Scholar] [CrossRef] [PubMed]
- Sobkowiak, L.; Bielewicz, D.; Malecka, E.M.; Jakobsen, I.; Albrechtsen, M.; Szweykowska-Kulinska, Z.; Pacak, A. The Role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front. Plant Sci. 2012, 3, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Rouached, H.; Secco, D.; Arpat, B.; Poirier, Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon Phosphate starvation in Arabidopsis. BMC Plant Biol. 2011, 11, 19. [Google Scholar] [CrossRef] [Green Version]
- Sega, P.; Kruszka, K.; Szewc, Ł.; Szweykowska-Kulińska, Z.; Pacak, A. Identification of transcription factors that bind to the 5′-UTR of the barley PHO2 gene. Plant Mol. Biol. 2019, 1–16. [Google Scholar] [CrossRef]
- Sun, L.; Song, L.; Zhang, Y.; Zheng, Z.; Liu, D. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to Phosphate starvation. Plant Physiol. 2016, 170, 499–514. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zheng, Z.; Song, L.; Liu, D. Functional characterization of Arabidopsis PHL4 in Plant response to Phosphate starvation. Front. Plant Sci. 2018, 9, 1432. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Miao, J.; Guo, J.; Shi, Z.; He, M.; Chen, Y.; Zhao, X.; Li, B.; Han, F.; et al. A Phosphate starvation response regulator Ta-PHR1 is involved in Phosphate signalling and increases grain yield in wheat. Ann. Bot. 2013, 111, 1139–1153. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Guo, Q.Q.; Chang, L.L.; Chen, L.; Zhao, C.Z.; Zhong, H.; Li, X.B. Brassica napus PHR1 gene encoding a MYB-like protein functions in response to Phosphate starvation. PLoS ONE 2012, 7, e44005. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Ruan, W.; Li, C.; Huang, F.; Zeng, M.; Liu, Y.; Yu, Y.; Ding, X.; Wu, Y.; Wu, Z.; et al. Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in Phosphate signaling and homeostasis in rice. Plant Physiol. 2015, 168, 1762–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, W.; Guo, M.; Wu, P.; Yi, K. Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol. Biol. 2017, 93, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Zhong, Y.; Wang, Y.; Wang, Z.; Zhang, L.; Shi, J.; Wu, Z.; Liu, Y.; Mao, C.; Yi, K.; et al. SPX4 negatively regulates Phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 2014, 26, 1586–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, A.E.; Grant, B.R.; Plaxton, W.C. Phosphite (phosphorous acid): Its relevance in the environment and agriculture and influence on plant Phosphate starvation response. J. Plant Nutr. 2001, 24, 1505–1519. [Google Scholar] [CrossRef]
- Decker, E.L.; Alder, A.; Hunn, S.; Ferguson, J.; Lehtonen, M.T.; Scheler, B.; Kerres, K.L.; Wiedemann, G.; Safavi-Rizi, V.; Nordzieke, S.; et al. Strigolactone biosynthesis is evolutionarily conserved, regulated by Phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 2017, 216, 455–468. [Google Scholar] [CrossRef] [Green Version]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Yang, Y.X.; Ahammed, G.J.; Wu, C.; Fan, S.Y.; Zhou, Y.H. Crosstalk among Jasmonate, Salicylate and Ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 2015, 16, 450–461. [Google Scholar] [CrossRef]
- Li, T.; Jia, K.P.; Lian, H.L.; Yang, X.; Li, L.; Yang, H.Q. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 454, 78–83. [Google Scholar] [CrossRef]
- Crombez, H.; Motte, H.; Beeckman, T. Tackling plant Phosphate starvation by the roots. Dev. Cell 2019, 48, 599–615. [Google Scholar] [CrossRef] [Green Version]
- Khan, G.A.; Vogiatzaki, E.; Glauser, G.; Poirier, Y. Phosphate deficiency induces the Jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016, 171, 632–644. [Google Scholar] [CrossRef] [Green Version]
- Castrillo, G.; Teixeira, P.J.; Paredes, S.H.; Law, T.F.; de Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, C.D.; et al. Root microbiota drive direct integration of Phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, F.; Shen, Y.; Zhan, Q.; Bai, B.; Chen, W.; Chi, Y. Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol. 2019, 19, 306. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Gerlach, N.; Sacristán, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramírez, D.; Bucher, M.; O’Connell, R.J.; et al. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are Phosphate status dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, D.K.; Chatterjee, S.; Datta, S.; Veer, V.; Walther, C. Role of Phosphate fertilizers in heavy metal uptake and detoxification of toxic metals. Chemosphere 2014, 108, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.F.; Rouached, H.; Tissot, N.; Gaymard, F.; Dubos, C. Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: Potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 2015, 6, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathan, K.K.; Amberger, A. Influence of iron on the uptake of phosphorus by maize. Plant Soil 1977, 46, 413–422. [Google Scholar] [CrossRef]
- Hirsch, J.; Marin, E.; Floriani, M.; Chiarenza, S.; Richaud, P.; Nussaume, L.; Thibaud, M.C. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 2006, 88, 1767–1771. [Google Scholar] [CrossRef]
- Bournier, M.; Tissot, N.; Mari, S.; Boucherez, J.; Lacombe, E.; Briat, J.F.; Gaymard, F. Arabidopsis ferritin 1 (AtFer1) gene regulation by the Phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and Phosphate homeostasis. J. Biol. Chem. 2013, 288, 22670–22680. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Hu, W.; Fan, X.; Chen, H.; Tang, M. Interactions between phosphorus, Zinc, and iron homeostasis in nonmycorrhizal and mycorrhizal plants. Front. Plant Sci. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Linn, J.; Ren, M.; Berkowitz, O.; Ding, W.; van der Merwe, M.J.; Whelan, J.; Jost, R. Root cell-specific regulators of Phosphate-dependent growth. Plant Physiol. 2017, 174, 1969–1989. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Luo, W.; Jain, A.; Liu, L.; Ai, H.; Liu, X.; Feng, B.; Zhang, L.; Zhang, Z.; Guohua, X.; et al. OsPHR3 affects the traits governing nitrogen homeostasis in rice. BMC Plant Biol. 2018, 18, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacak, A.; Barciszewska-Pacak, M.; Świda-Barteczka, A.; Kruszka, K.; Sega, P.; Milanowska, K.; Jakobsen, I.; Jarmołowski, A.; Szweykowska-Kulińska, Z. Heat stress affects pi-related genes expression and inorganic Phosphate deposition/accumulation in Barley. Front. Plant Sci. 2016, 7, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Ding, L.; Casola, C.; Ripoll, D.R.; Feschotte, C.; Wang, H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 2007, 318, 1302–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 2017, 29, 2269–2284. [Google Scholar] [CrossRef] [Green Version]
- Lei, M.; Zhu, C.; Liu, Y.; Karthikeyan, A.S.; Bressan, R.A.; Raghothama, K.G.; Liu, D. Ethylene signalling is involved in regulation of Phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 2011, 189, 1084–1095. [Google Scholar] [CrossRef]
- Thibaud, M.C.; Arrighi, J.F.; Bayle, V.; Chiarenza, S.; Creff, A.; Bustos, R.; Paz-Ares, J.; Poirier, Y.; Nussaume, L. Dissection of local and systemic transcriptional responses to Phosphate starvation in Arabidopsis. Plant J. 2010, 64, 775–789. [Google Scholar] [CrossRef]
- Neumann, G. The role of ethylene in plant adaptations for Phosphate acquisition in soils—A review. Front. Plant Sci. 2016, 6, 1224. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, V.K.; Smith, A.P. Ethylene’s role in Phosphate starvation signaling: More than just a root growth regulator. Plant Cell Physiol. 2012, 53, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.L.; Ma, G.J.; Zhang, M.L.; Xiong, H.; Wu, H.; Zhao, C.Z.; Liu, C.S.; Jia, H.X.; Chen, L.; Kjorven, J.O.; et al. The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol. 2018, 178, 413–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzucotelli, E.; Mastrangelo, A.M.; Crosatti, C.; Guerra, D.; Stanca, A.M.; Cattivelli, L. Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008, 174, 420–431. [Google Scholar] [CrossRef]
- Praefcke, G.J.; Hofmann, K.; Dohmen, R.J. SUMO playing tag with ubiquitin. Trends Biochem. Sci. 2012, 37, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Rus, A.; Sharkhuu, A.; Yokoi, S.; Karthikeyan, A.S.; Raghothama, K.G.; Baek, D.; Koo, Y.D.; Jin, J.B.; Bressan, R.A.; et al. The Arabidopsis SUMO E3 ligase SIZ1 controls Phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 2005, 102, 7760–7765. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sun, R.; Cao, Y.; Pei, W.; Sun, Y.; Zhou, H.; Wu, X.; Zhang, F.; Luo, L.; Shen, Q.; et al. OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to Phosphate and nitrogen in rice. Plant Cell Physiol. 2015, 56, 2381–2395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.F.; Zhou, L.J.; Li, Y.Y.; You, C.X.; Sha, G.L.; Hao, Y.J. Apple SUMO E3 ligase MdSIZ1 is involved in the response to Phosphate deficiency. J. Plant Physiol. 2019, 232, 216–225. [Google Scholar] [CrossRef]
- Park, H.C.; Kim, H.; Koo, S.C.; Park, H.J.; Cheong, M.S.; Hong, H.; Baek, D.; Chung, W.S.; Kim, D.H.; Bressan, R.A.; et al. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant. Cell Environ. 2010, 33, 1923–1934. [Google Scholar] [CrossRef]
- Jin, J.B.; Jin, Y.H.; Lee, J.; Miura, K.; Yoo, C.Y.; Kim, W.Y.; Van Oosten, M.; Hyun, Y.; Somers, D.E.; Lee, I.; et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008, 53, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.J.; Zhang, C.L.; Zhang, R.F.; Wang, G.L.; Li, Y.Y.; Hao, Y.J. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+ -ATPase activity and iron homeostasis. Plant Physiol. 2019, 179, 88–106. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.; Lin, X.L.; Kong, X.; Qu, G.P.; Cai, B.; Lee, J.; Jin, J.B. SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol. Plant 2019, 12, 215–228. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.; Srivastava, A.P.; Esmaeili, N.; Hu, W.; Shen, G. Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought-, heat-, and salt-tolerance simultaneously. PLoS ONE 2018, 13, e0201716. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.H. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the Phosphate transporter PT2 for degradation during the regulation of Arabidopsis Phosphate homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane–localized Phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 integrates nitrate and Phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.; Khan, S.; Fahad, S.; Faisal, S.; Hussain, S.; Ali, S.; Ali, A. Effect of different levels of nitrogen and phosphorus on the phenology and yield of maize varieties. Am. J. Plant Sci. 2014, 5, 2582–2590. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Mazer, S.J.; Guo, H.; Zhang, N.; Weiner, J.; Hu, S. Nitrogen: Phosphorous supply ratio and allometry in five alpine plant species. Ecol. Evol. 2016, 6, 8881–8892. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ying, S.; Huang, H.; Li, K.; Wu, P.; Shou, H. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009, 57, 895–904. [Google Scholar] [CrossRef]
- Ruan, W.; Guo, M.; Wang, X.; Guo, Z.; Xu, Z.; Xu, L.; Zhao, H.; Sun, H.; Yan, C.; Yi, K. Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal Phosphate sensor, for Phosphate homeostasis and signaling in rice. Mol. Plant 2019, 12, 1060–1074. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; de Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodriguez, J.; et al. SPX1 is a Phosphate-dependent inhibitor of Phosphate starvation response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [Green Version]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef]
- Li, Y.; Wu, H.; Fan, H.; Zhao, T.; Ling, H.Q. Characterization of the AtSPX3 promoter elucidates its complex regulation in response to phosphorus deficiency. Plant Cell Physiol. 2016, 57, 1767–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, M.B.; Ng, S.; Berkowitz, O.; De Clercq, I.; Mao, C.; Shou, H.; Whelan, J.; Jost, R. SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 2019, 181, 332–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Wang, Y.; Guo, J.; Zhu, X.; Shi, J.; He, Q.; Liu, Y.; Wu, Y.; Zhang, L.; Lv, Q.; et al. Rice SPX6 negatively regulates the Phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 2018, 219, 135–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jiang, Z.; Wang, W.; Qiu, Y.; Zhang, Z.; Liu, Y.; Li, A.; Gao, X.; Liu, L.; Qian, Y.; et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 2019, 5, 401–413. [Google Scholar] [CrossRef]
- Qi, W.; Manfield, I.W.; Muench, S.P.; Baker, A. AtSPX1 affects the AtPHR1–DNA-binding equilibrium by binding monomeric AtPHR1 in solution. Biochem. J. 2017, 474, 3675–3687. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y.; et al. Rice SPX1 and SPX2 inhibit Phosphate starvation responses through interacting with PHR2 in a Phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, A. Role of Ca2+ and EGTA on stomatal movements in Commelina communis L. Plant Physiol. 1985, 79, 1003–1005. [Google Scholar] [CrossRef] [Green Version]
- Tai, L.; Li, B.B.; Nie, X.M.; Zhang, P.P.; Hu, C.H.; Zhang, L.; Liu, W.T.; Li, W.Q.; Chen, K.M. Calmodulin is the fundamental regulator of NADK-mediated NAD signaling in plants. Front. Plant Sci. 2019, 10, 681. [Google Scholar] [CrossRef] [Green Version]
- Yoo, C.Y.; Mano, N.; Finkler, A.; Weng, H.; Day, I.S.; Reddy, A.S.N.; Poovaiah, B.W.; Fromm, H.; Hasegawa, P.M.; Mickelbart, M.V. A Ca2+/CaM-regulated transcriptional switch modulates stomatal development in response to water deficit. Sci. Rep. 2019, 9, 12282. [Google Scholar] [CrossRef] [Green Version]
- Allen, G.J.; Muir, S.R.; Sanders, D. Release of Ca2+ from individual plant vacuoles by both InsP3 and cyclic ADP-ribose. Science 1995, 268, 735–737. [Google Scholar] [CrossRef]
- Shears, S.B.; Ganapathi, S.B.; Gokhale, N.A.; Schenk, T.M.; Wang, H.; Weaver, J.D.; Zaremba, A.; Zhou, Y. Defining signal transduction by inositol Phosphates. Subcell. Biochem. 2012, 59, 389–412. [Google Scholar] [PubMed] [Green Version]
- Hermosura, M.C.; Takeuchi, H.; Fleig, A.; Riley, A.M.; Potter, B.V.; Hirata, M.; Penner, R. InsP4 facilitates store-operated calcium influx by inhibition of InsP3 5-phosphatase. Nature 2000, 408, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Menniti, F.S.; Oliver, K.G.; Putney, J.W.; Shears, S.B. Inositol Phosphates and cell signaling: New views of InsP5 and InsP6. Trends Biochem. Sci. 1993, 18, 53–56. [Google Scholar] [CrossRef]
- Kuo, H.F.; Chang, T.Y.; Chiang, S.F.; Wang, W.D.; Charng, Y.Y.; Chiou, T.J. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates Phosphate homeostasis at the transcriptional level. Plant J. 2014, 80, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.F.; Hsu, Y.Y.; Lin, W.C.; Chen, K.Y.; Munnik, T.; Brearley, C.A.; Chiou, T.J. Arabidopsis inositol Phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining Phosphate homeostasis. Plant J. 2018, 95, 613–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, R.; Gerasimaite, R.; Jung, J.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic Phosphate homeostasis by inositol Polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Ma, G.; Sui, L.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.E.; Wittwer, C.; et al. Inositol Pyrophosphate InsP8 acts as an intracellular Phosphate signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. [Google Scholar] [CrossRef]
- Shears, S.B. Inositol pyrophosphates: Why so many Phosphates? Adv. Biol. Regul. 2015, 57, 203–216. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol Pyrophosphate kinases/phosphatases control plant Phosphate homeostasis. eLife 2019, 8, e43582. [Google Scholar] [CrossRef]
- Wang, H.; Nair, V.S.; Holland, A.A.; Capolicchio, S.; Jessen, H.J.; Johnson, M.K.; Shears, S.B. Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S]2+ cluster which inhibits inositol Pyrophosphate 1-phosphatase activity. Biochemistry 2015, 54, 6462–6474. [Google Scholar] [CrossRef] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by Inositol-Phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Laha, D.; Johnen, P.; Azevedo, C.; Dynowski, M.; Weiß, M.; Capolicchio, S.; Mao, H.; Iven, T.; Steenbergen, M.; Freyer, M.; et al. VIH2 regulates the synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-dependent defenses in Arabidopsis. Plant Cell 2015, 27, 1082–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376. [Google Scholar] [CrossRef] [Green Version]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of molecular integrators shows that nitrogen actively controls the Phosphate starvation response in plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef]
- Huang, T.K.; Han, C.L.; Lin, S.I.; Chen, Y.J.; Tsai, Y.C.; Chen, Y.R.; Chen, J.W.; Lin, W.Y.; Chen, P.M.; Liu, T.Y.; et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sega, P.; Pacak, A. Plant PHR Transcription Factors: Put on A Map. Genes 2019, 10, 1018. https://doi.org/10.3390/genes10121018
Sega P, Pacak A. Plant PHR Transcription Factors: Put on A Map. Genes. 2019; 10(12):1018. https://doi.org/10.3390/genes10121018
Chicago/Turabian StyleSega, Paweł, and Andrzej Pacak. 2019. "Plant PHR Transcription Factors: Put on A Map" Genes 10, no. 12: 1018. https://doi.org/10.3390/genes10121018
APA StyleSega, P., & Pacak, A. (2019). Plant PHR Transcription Factors: Put on A Map. Genes, 10(12), 1018. https://doi.org/10.3390/genes10121018




