Transcriptomic Profiling Identifies Candidate Genes Involved in the Salt Tolerance of the Xerophyte Pugionium cornutum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions and Salt Treatments in Sand Culture Experiments
2.2. Determination of the Physiological Indices Related to Plant Growth, Stability of the Cell Membrane, and Photosynthesis
2.3. Plant Growth Conditions and Treatments for Transcriptome Sequencing
2.4. Transcriptome Sequencing
2.5. Differentially Expressed Genes Analysis
2.6. Real-Time Quantitative PCR Validation of DEG Results
3. Results
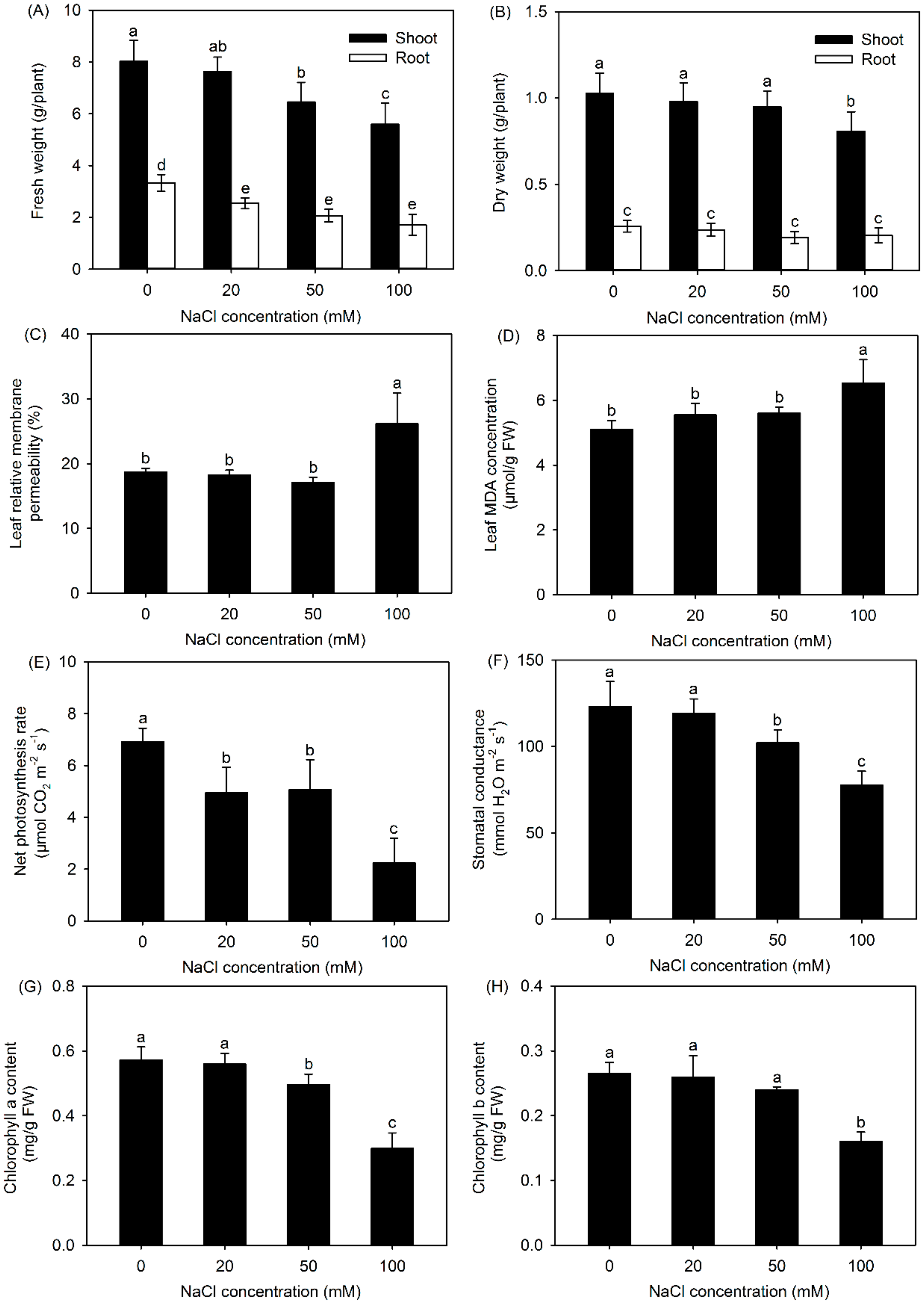
3.1. Changes in Plant Growth, Cell Membrane Stability, and Photosynthesis Performance of P. cornutum under Different NaCl Treatments
3.2. Transcriptome Sequencing, De Novo Assembly, and Functional Annotation
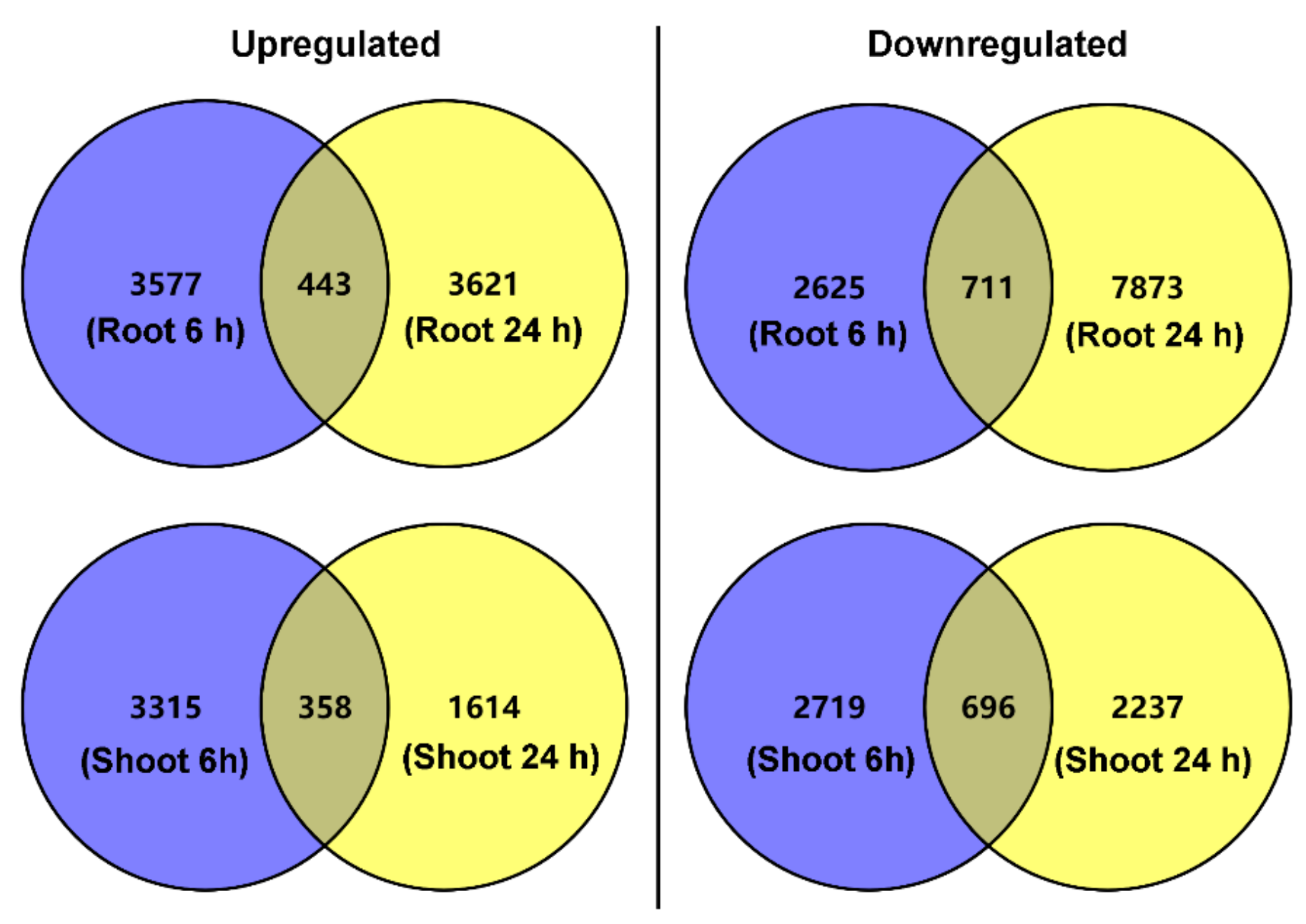
3.3. Identification of DEGs in the Roots and Shoots of P. cornutum Treated with 50 mM NaCl
3.4. DEGs Related to Ion Transport
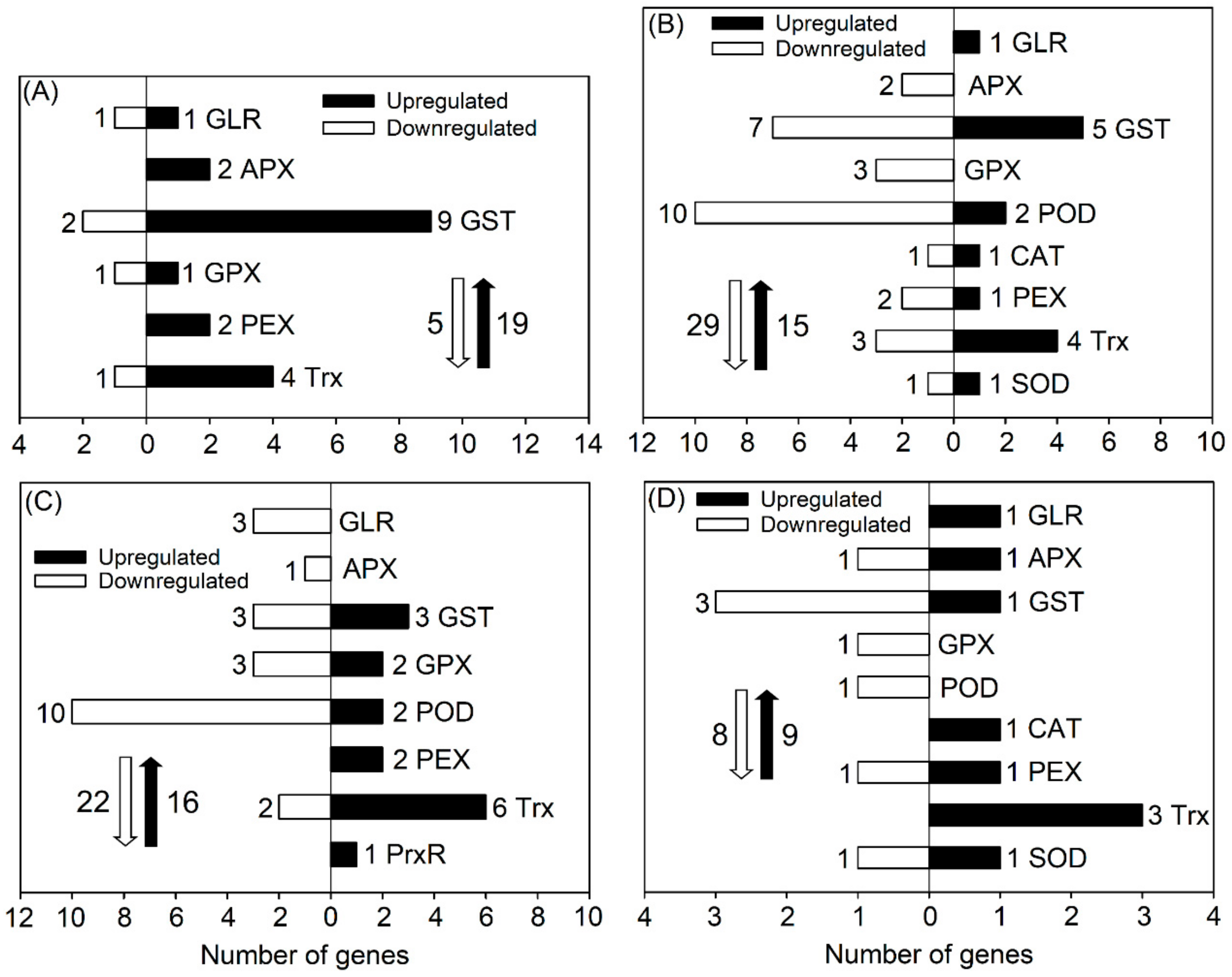
3.5. DEGs Related to the ROS-Scavenging System
3.6. DEGs Related to Photosynthesis
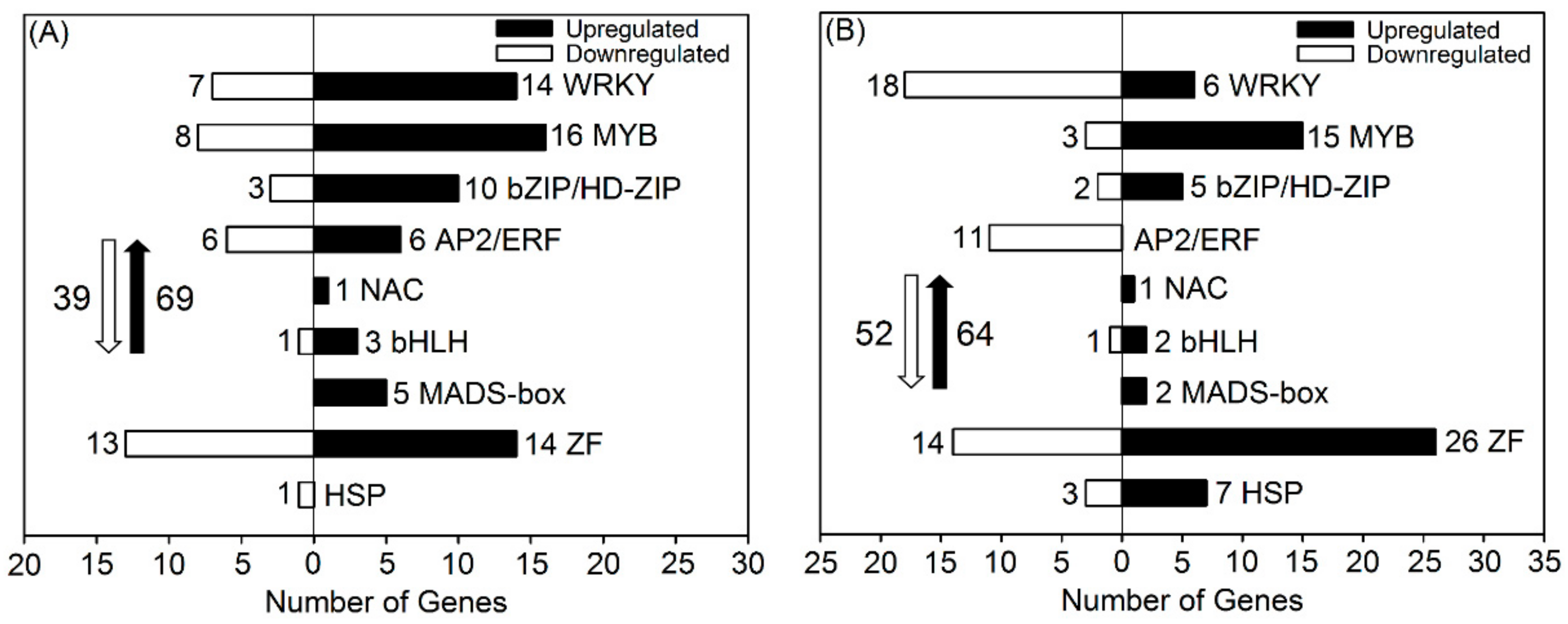
3.7. DEGs Related to Transcription Factors
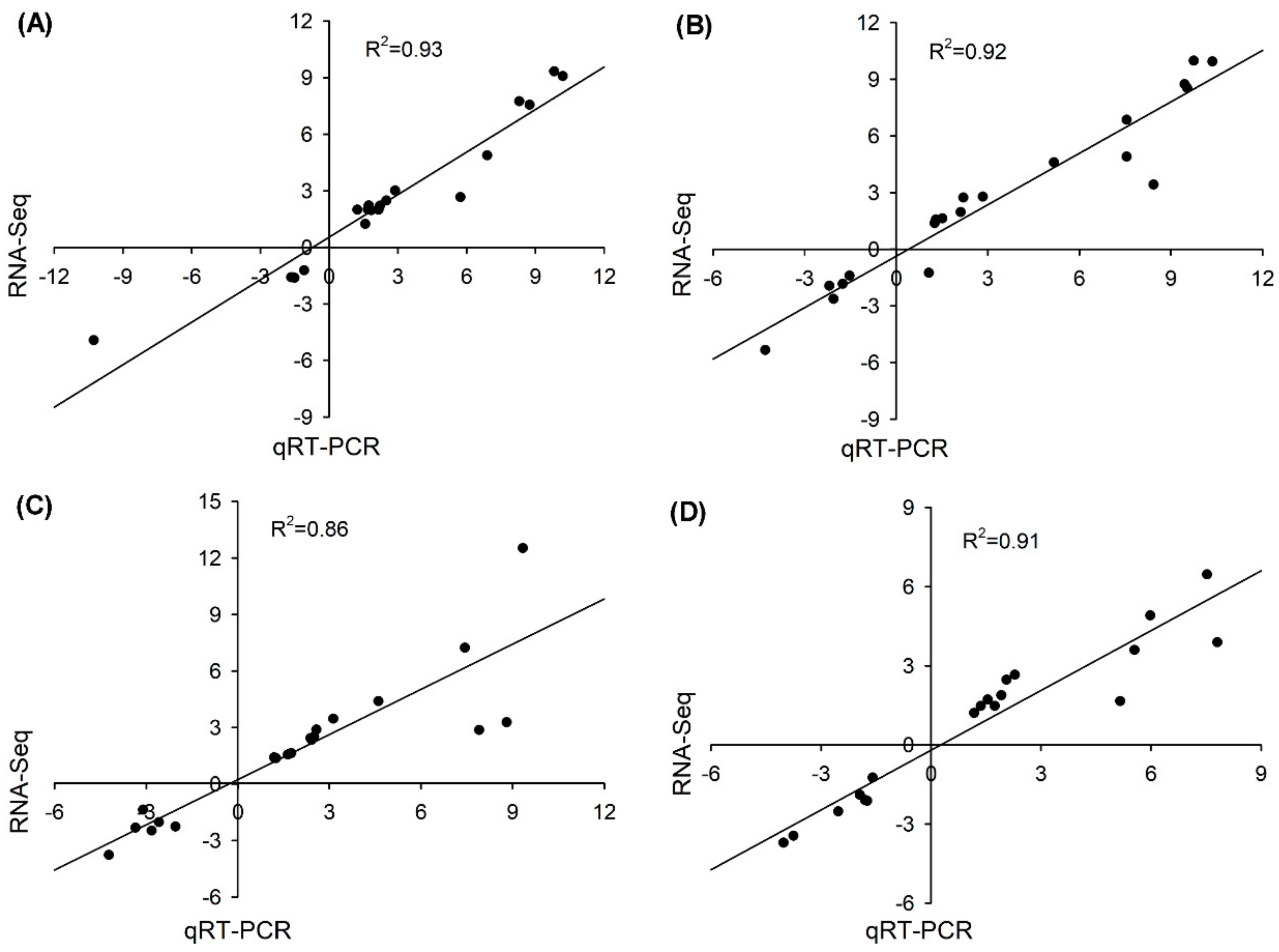
3.8. Validation of the RNA-seq Results
4. Discussion
4.1. The Possible Molecular Basis Underlying Cl− Transport in the Cl−-Accumulating Species P. cornutum
4.2. The Transport of Other Important Ions Plays a Vital Role in the Response to Salt Stress in P. cornutum
4.3. The ROS-Scavenging System is Important for Salt Stress Adaptation in P. cornutum
4.4. High Efficiency for Stomatal Movement and Carbon Fixation is Essential for the Adaptation of P. Cornutum to Salt Stress
4.5. Transcription Factors Play Important Roles in Regulating Salt-Responsive Genes in P. Cornutum under Salt Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Bose, J.; Fuglsang, A.T.; Pottosin, I. On a quest for stress tolerance genes: Membrane transporters in sensing and adapting to hostile soils. J. Exp. Bot. 2016, 67, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.H.; Pieckenstain, F.L.; Szymanski, J.; Erban, A.; Bromke, M.; Hannah, M.A.; Kraemer, U.; Kopka, J.; Udvardi, M.K. Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS ONE 2011, 6, 1–11. [Google Scholar] [CrossRef]
- Ma, Q.; Bao, A.K.; Chai, W.W.; Wang, W.Y.; Zhang, J.L.; Li, Y.X.; Wang, S.M. Transcriptomic analysis of the succulent xerophyte Zygophyllum xanthoxylum in response to salt treatment and osmotic stress. Plant Soil 2016, 402, 343–361. [Google Scholar] [CrossRef]
- Yu, Q.S.; Wang, Q.; Wang, A.L.; Wu, G.L.; Liu, J.Q. Interspecific delimitation and phylogenetic origin of Pugionium (Brassicaceae). J. Syst. Evol. 2010, 48, 195–206. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Zhang, C.; Chen, B.; Hui, L.; Shen, Y. Compositional and gastrointestinal prokinetic studies of Pugionium (L.). Food Chem. 2015, 186, 285–291. [Google Scholar] [CrossRef]
- Li, N.; Hao, L.Z.; Song, Z.W.; Zhang, F.L.; Yang, Z.R.; Wen, J. Effect of NaCl on germination and seedlings establishment in Pugionium cornutum (L.) Gaertn. J. Inner Mong. Agri. Univ. 2010, 4, 58–63. [Google Scholar]
- Pang, J.; Hao, L.Z.; Zhang, F.L.; Zhao, P.; Yang, Z.R. The response of active oxygen species and ascorbic acid in Pugionium cornutum (L.) Gaertn. leaves to drought stress. Plant Physiol. J. 2013, 49, 57–62. [Google Scholar] [CrossRef]
- Yue, L.J.; Cui, Y.N.; Yuan, K.; Kang, J.J.; Wang, S.M. The osmotic adjustment in Pugionium cornutum subjected to salt stress. Plant Physiol. J. 2016, 52, 569–574. [Google Scholar] [CrossRef]
- Yue, L.J.; Yuan, K.; Li, H.W.; Kang, J.J.; Wang, S.M. Adaptive responses of eremophyte Pugionium cornutum seedlings to different concentrations of NaCl. Acta. Pratacult. Sin. 2016, 25, 144–152. [Google Scholar] [CrossRef]
- Li, B.; Tester, M.; Gilliham, M. Chloride on the move. Trends Plant Sci. 2017, 22, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods. 2008, 5, 16–18. [Google Scholar] [CrossRef]
- Yue, L.J.; Li, S.X.; Ma, Q.; Zhou, X.R.; Wu, G.Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. NaCl stimulates growth and alleviates water stress in the xerophyte Zygophyllum xanthoxylum. J. Arid Environ. 2012, 87, 153–160. [Google Scholar] [CrossRef]
- Ma, Q.; Yue, L.J.; Zhang, J.L.; Wu, G.Q.; Bao, A.K.; Wang, S.M. Sodium chloride improves photosynthesis and water status in the succulent xerophyte Zygophyllum xanthoxylum. Tree Physiol. 2012, 32, 4–13. [Google Scholar] [CrossRef]
- Dang, Z.H.; Zheng, L.L.; Wang, J.; Gao, Z.; Wu, S.B.; Qi, Z.; Wang, Y.C. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genom. 2013, 14, 29. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, L.; Cui, Y.N.; Wang, S.M.; Bao, A.K. Identification of candidate genes related to salt tolerance of the secretohalophyte Atriplex canescens by transcriptomic analysis. BMC Plant Biol. 2019, 19, 213. [Google Scholar] [CrossRef]
- Sairam, R.K.; Saxena, D.C. Oxidative stress and antioxidants in wheat genotypes: Possible mechanism of water stress tolerance. J. Agron. Crop Sci. 2000, 184, 55–61. [Google Scholar] [CrossRef]
- Ali, A.; Raddat, N.; Aman, R.; Kim, S.; Park, H.C.; Jan, M.; Baek, D.; Khan, I.U.; Oh, D.H.; Lee, S.Y.; et al. A single amino acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol. 2016, 171, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Siahpoosh, M.R.; Sanchez, D.H.; Schlereth, A.; Schlereth, A.; Scofield, G.N.; Furbank, R.T.; van Dongen, J.T.; Kopka, J. Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309. Plant Sci. 2012, 182, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Gaxiola, R.A.; Palmgren, M.G.; Schumacher, K. Plant proton pumps. FEBS Lett. 2007, 581, 2204–2214. [Google Scholar] [CrossRef] [PubMed]
- Nevo, R.; Charuvi, D.; Tsabari, O.; Reich, Z. Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 2012, 70, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Broadley, M.R. Chloride in soils and its uptake and movement within the plant: A review. Ann. Bot. 2001, 88, 967–988. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Skerrett, I.M. Root ion channels and salinity. Sci. Hortic. 1999, 78, 175–235. [Google Scholar] [CrossRef]
- Wen, Z.; Tyerman, S.D.; Dechorgnat, J.; Ovchinnikova, E.; Dhugga, K.S.; Kaiser, B.N. Maize NPF6 proteins are homologs of Arabidopsis chl1 that are selective for both nitrate and chloride. Plant Cell 2017, 29, 2581–2596. [Google Scholar] [CrossRef]
- Qiu, J.; Henderson, S.W.; Tester, M.; Roy, S.J.; Gilliham, M. SLAH1, a homologue of the slow type anion channel SLAC1, modulates shoot Cl- accumulation and salt tolerance in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 4495–4505. [Google Scholar] [CrossRef]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 2010, 33, 566–589. [Google Scholar] [CrossRef] [PubMed]
- Wegner, L.H. Root pressure and beyond: Energetically uphill water transport into xylem vessels? J. Exp. Bot. 2014, 65, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.T.; Agorio, A.; Jossier, M.; Depré, S.; Thomine, S.; Filleur, S. Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.P.; Wang, L.; Liu, A.; Yu, B.; Lam, H.M. GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 2016, 7, 1082. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu Rev Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, M.L.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef]
- Chen, C.Z.; Lv, X.F.; Li, J.Y.; Yi, H.Y.; Gong, J.M. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 2012, 159, 1582–1590. [Google Scholar] [CrossRef]
- Gobert, A.; Park, G.; Amtmann, A.; Sanders, D.; Maathuis, F.J. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a nonselective ion transporter involved in germination and cation transport. J. Exp. Bot. 2006, 57, 791–800. [Google Scholar] [CrossRef]
- Qi, Z.; Stephens, N.R.; Spalding, E.P. Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 2006, 142, 963–971. [Google Scholar] [CrossRef]
- Wang, P.; Li, Z.; Wei, J.; Zhao, Z.; Sun, D.; Cui, S. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J. Biol. Chem. 2012, 287, 44062–44070. [Google Scholar] [CrossRef]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Z.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.H.; Luan, S. AtKUP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 1998, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Apse, M.P. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, Y.X.; Yuan, H.J.; Hu, J.; Wei, L.; Bao, A.K.; Zhang, J.L.; Wang, S.M. ZxSOS1 is essential for long-distance transport and spatial distribution of Na+ and K+ in the xerophyte Zygophyllum xanthoxylum. Plant Soil 2014, 374, 661–676. [Google Scholar] [CrossRef]
- Yuan, H.J.; Ma, Q.; Wu, G.Q.; Wang, P.; Hu, J.; Wang, S.M. ZxNHX controls Na+ and K+ homeostasis at the whole-plant level in Zygophyllum xanthoxylum through feedback regulation of the expression of genes involved in their transport. Ann. Bot. 2015, 115, 495–507. [Google Scholar] [CrossRef]
- Bassil, E.; Ohto, M.; Esumi, T.; Tajima, H.; Zhu, Z.; Cagnac, O.; Belmonte, M.; Peleg, Z.; Yamaguchi, T.; Blumwald, E. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 2011, 23, 224–239. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef]
- Roelfsema, M.R.; Hedrich, R. In the light of stomatal opening: New insights into “the watergate”. New Phytol. 2010, 167, 665–691. [Google Scholar] [CrossRef]
- Hosy, E.; Vavasseur, A.; Mouline, K.; Dreyer, I.; Gaymard, F.; Porée, F.; Boucherez, J.; Lebaudy, A.; Bouchez, D.; Véry, A.A.; et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA 2003, 100, 5549–5554. [Google Scholar] [CrossRef]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.S.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Kroniewicz, L.; Dalmas, F.; Le Thiec, D.; Ephritikhine, G.; Barbier-Brygoo, H.; Vavasseur, A.; Filleur, S.; Leonhardt, N. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 2010, 64, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Mori, I.C.; Furuichi, T.; Munemasa, S.; Toyooka, K.; Matsuoka, K.; Murata, Y.; Yamamoto, Y. Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 2010, 51, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Deyholos, M.K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef]


| Unigenes | Total Number | Total Length (bp) | Mean Length (bp) |
|---|---|---|---|
| Shoots | 64,978 | 70,865,211 | 1091 |
| Roots | 80,307 | 80,091,654 | 997 |
| All | 72,086 | 89,575,317 | 1243 |
| Gene ID | Gene Name | Function |
|---|---|---|
| CL5647.Contig1_All | SLAH1 | Cl− transport from roots to shoots |
| CL2477.Contig4_All | CLCg | Cl− vacuole compartmentalization |
| CL7387.Contig1_All | NPF6.4 | Cl− uptake at root epidermis |
| CL1096.Contig4_All | NHX5 | Na+/H+ antiporter |
| CL1096.Contig10_All | NHX6 | Na+/H+ antiporter |
| CL3604.Contig8_All | GLR3.3 | Cation channel activator |
| CL3206.Contig3_All | Sultr1.2 | SO42− uptake |
| CL7785.Contig1_All | Sultr3.3 | SO42− transporter |
| CL564.Contig8_All | PHT4.3 | PO43− transporter |
| CL2740.Contig1_All | PDR2 | Mn2+ transporter |
| CL666.Contig1_All | BOR4 | BO33− exporting from cytoplasm |
| Gene ID | Gene Name | Function |
|---|---|---|
| CL4002.Contig2_All | CCC1 | Cl− retrieval from xylem sap |
| CL2577.Contig2_All | CLCf | Cl− channel |
| CL2477.Contig4_All | CLCg | Cl− vacuole compartmentalization |
| Unigene7084_All | NPF5.2 | Cl− and/or NO3− transporter |
| CL3282.Contig2_All | NPF6.2 | Cl− and/or NO3− transporter |
| CL6278.Contig1_All | NPF8.1 | Cl− and/or NO3− transporter |
| CL2487.Contig3_All | NPF5.9 | Cl− and/or NO3− transporter |
| CL1096.Contig4_All | NHX5 | Na+/H+ antiporter |
| CL1096.Contig10_All | NHX6 | Na+/H+ antiporter |
| CL3834.Contig2_All | HKT1 | Na+ and/or K+ cellular efflux |
| CL1280.Contig5_All | KUP5 | K+ transporter |
| Unigene6900_All | CNGC1 | Ca2+ root uptake |
| Unigene13264_All | CTR5 | Cu2+ transporter |
| Unigene1617_All | MGT10 | Mg2+ transporter |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.-N.; Wang, F.-Z.; Yang, C.-H.; Yuan, J.-Z.; Guo, H.; Zhang, J.-L.; Wang, S.-M.; Ma, Q. Transcriptomic Profiling Identifies Candidate Genes Involved in the Salt Tolerance of the Xerophyte Pugionium cornutum. Genes 2019, 10, 1039. https://doi.org/10.3390/genes10121039
Cui Y-N, Wang F-Z, Yang C-H, Yuan J-Z, Guo H, Zhang J-L, Wang S-M, Ma Q. Transcriptomic Profiling Identifies Candidate Genes Involved in the Salt Tolerance of the Xerophyte Pugionium cornutum. Genes. 2019; 10(12):1039. https://doi.org/10.3390/genes10121039
Chicago/Turabian StyleCui, Yan-Nong, Fang-Zhen Wang, Cheng-Hang Yang, Jian-Zhen Yuan, Huan Guo, Jin-Lin Zhang, Suo-Min Wang, and Qing Ma. 2019. "Transcriptomic Profiling Identifies Candidate Genes Involved in the Salt Tolerance of the Xerophyte Pugionium cornutum" Genes 10, no. 12: 1039. https://doi.org/10.3390/genes10121039
APA StyleCui, Y.-N., Wang, F.-Z., Yang, C.-H., Yuan, J.-Z., Guo, H., Zhang, J.-L., Wang, S.-M., & Ma, Q. (2019). Transcriptomic Profiling Identifies Candidate Genes Involved in the Salt Tolerance of the Xerophyte Pugionium cornutum. Genes, 10(12), 1039. https://doi.org/10.3390/genes10121039





