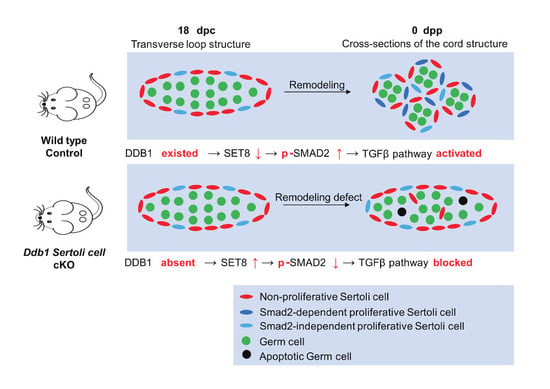
DDB1 Regulates Sertoli Cell Proliferation and Testis Cord Remodeling by TGFβ Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Mice
2.2. Sertoli Cell Isolation
2.3. Western Blotting
2.4. Histological Analysis
2.5. BrdU Labeling
2.6. Hematoxylin and Eosin (H&E) Staining and Immunostaining
2.7. Statistical Analysis
3. Results
3.1. DDB1 Expression and Localization in Testes
3.2. Genetic Deletion of Ddb1 in Sertoli Cells
3.3. Disturbed Morphogenesis of Testis Cords in Neonatal Ddb1 cKO Mice
3.4. Germ Cell Accumulation and Decreased Sertoli Cells in Neonatal Ddb1 cKO Mice
3.5. Sertoli Cell Proliferation Decreased in Neonatal Ddb1 cKO Mice
3.6. Variable Expression of Proteins Associated with DDB1 in Ddb1 cKO Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, S.-R.; Liu, Y.-X. Testis Cord Maintenance in Mouse Embryos: Genes and Signaling. Boil. Reprod. 2016, 94, 42. [Google Scholar] [CrossRef] [PubMed]
- Cool, J.; DeFalco, T.; Capel, B. Testis formation in the fetal mouse: Dynamic and complex de novo tubulogenesis. Wiley Interdiscip. Rev. Dev. Boil. 2012, 1, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Brennan, J.; Capel, B. One tissue, two fates: Molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 2004, 5, 509. [Google Scholar] [CrossRef] [PubMed]
- Combes, A.N.; Lesieur, E.; Harley, V.R.; Sinclair, A.H.; Little, M.H.; Wilhelm, D.; Koopman, P. Three-dimensional visualization of testis cord morphogenesis, a novel tubulogenic mechanism in development. Dev. Dyn. 2009, 238, 1033–1041. [Google Scholar] [CrossRef]
- Svingen, T.; Koopman, P. Building the mammalian testis: Origins, differentiation, and assembly of the component cell populations. Genes Dev. 2013, 27, 2409–2426. [Google Scholar] [CrossRef]
- Das, D.K.; Sanghavi, D.; Gawde, H.; Idicula-Thomas, S.; Vasudevan, L. Novel homozygous mutations in Desert hedgehog gene in patients with 46,XY complete gonadal dysgenesis and prediction of its structural and functional implications by computational methods. Eur. J. Med Genet. 2011, 54, e529–e534. [Google Scholar] [CrossRef]
- Angelopoulou, R.; Balla, M.; Lavranos, G.; Chalikias, M.; Kitsos, C.; Baka, S.; Kittas, C. Sertoli cell proliferation in the fetal and neonatal rat testis: A continuous phenomenon? Acta Histochem. 2008, 110, 341–347. [Google Scholar] [CrossRef]
- Archambeault, D.R.; Yao, H.H.-C. Activin A, a product of fetal Leydig cells, is a unique paracrine regulator of Sertoli cell proliferation and fetal testis cord expansion. Proc. Natl. Acad. Sci. USA 2010, 107, 10526–10531. [Google Scholar] [CrossRef]
- Archambeault, D.R.; Yao, H.H.-C. Loss of smad4 in Sertoli and Leydig cells leads to testicular dysgenesis and hemorrhagic tumor formation in mice. Boil. Reprod. 2014, 90, 62. [Google Scholar] [CrossRef]
- Memon, M.A.; Anway, M.D.; Covert, T.R.; Uzumcu, M.; Skinner, M.K. Transforming growth factor beta (TGFbeta1, TGFbeta2 and TGFbeta3) null-mutant phenotypes in embryonic gonadal development. Mol. Cell. Endocrinol. 2008, 294, 70–80. [Google Scholar] [CrossRef]
- Miles, D.C.; Wakeling, S.I.; Stringer, J.M.; van den Bergen, J.A.; Wilhelm, D.; Sinclair, A.H.; Western, P.S. Signaling through the TGF beta-activin receptors ALK4/5/7 regulates testis formation and male germ cell development. PLoS ONE 2013, 8, e54606. [Google Scholar] [CrossRef] [PubMed]
- Cang, Y.; Zhang, J.; Nicholas, S.A.; Kim, A.L.; Zhou, P.; Goff, S.P. DDB1 is essential for genomic stability in developing epidermis. Proc. Natl. Acad. Sci. USA 2007, 104, 2733–2737. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, C.; Fleck, O.; Hansen, H.A.; Liu, C.; Slaaby, R.; Carr, A.M.; Nielsen, O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005, 19, 853–862. [Google Scholar] [CrossRef]
- Jackson, S.; Xiong, Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009, 34, 562–570. [Google Scholar] [CrossRef]
- Lee, J.; Zhou, P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 2007, 26, 775–780. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q.E.; Zhu, Q.; El-Mahdy, M.A.; Wani, G.; Praetorius-Ibba, M.; Wani, A.A. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006, 66, 8590–8597. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, Y.L.; Pan, W.W.; Li, X.M.; Wang, Z.W.; Ge, Z.J.; Zhou, J.J.; Cang, Y.; Tong, C.; Sun, Q.Y.; et al. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science 2013, 342, 1518–1521. [Google Scholar] [CrossRef]
- Yu, J.; Lan, X.; Chen, X.; Yu, C.; Xu, Y.; Liu, Y.; Xu, L.; Fan, H.Y.; Tong, C. Protein synthesis and degradation are essential to regulate germline stem cell homeostasis in Drosophila testes. Development 2016, 143, 2930–2945. [Google Scholar] [CrossRef]
- Cang, Y.; Zhang, J.; Nicholas, S.A.; Bastien, J.; Li, B.; Zhou, P.; Goff, S.P. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 2006, 127, 929–940. [Google Scholar] [CrossRef]
- Lécureuil, C.; Fontaine, I.; Crepieux, P.; Guillou, F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 2002, 33, 114–118. [Google Scholar] [CrossRef]
- Jiang, H.; Gao, Q.; Zheng, W.; Yin, S.; Wang, L.; Zhong, L.; Ali, A.; Khan, T.; Hao, Q.; Fang, H. MOF influences meiotic expansion of H2AX phosphorylation and spermatogenesis in mice. PLoS Genet. 2018, 14, e1007300. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ma, T.; Zhang, Y.; Zhang, H.; Yin, S.; Zheng, W.; Wang, L.; Wang, Z.; Khan, M.; Sheikh, S.W.; et al. Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol. Reprod. 2015, 92, 79. [Google Scholar] [CrossRef] [PubMed]
- Vergouwen, R.; Jacobs, S.; Huiskamp, R.; Davids, J.; De Rooij, D. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. Reproduction 1991, 93, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, T.M. A review of recent advances in stereology for quantifying neural structure. J. Neurocytol. 1992, 21, 313–328. [Google Scholar] [CrossRef]
- Wreford, N.G. Theory and practice of stereological techniques applied to the estimation of cell number and nuclear volume in the testis. Microsc. Res. Tech. 1995, 32, 423–436. [Google Scholar] [CrossRef]
- Rebourcet, D.; Darbey, A.; Monteiro, A.; Soffientini, U.; Tsai, Y.T.; Handel, I.; Pitetti, J.-L.; Nef, S.; Smith, L.B.; O’Shaughnessy, P.J. Sertoli cell number defines and predicts germ and Leydig cell population sizes in the adult mouse testis. Endocrinology 2017, 158, 2955–2969. [Google Scholar] [CrossRef]
- Russell, L.D.; Ettlin, R.A.; Hikim, A.P.S.; Clegg, E.D. Histological and histopathological evaluation of the testis. Int. J. Androl. 1993, 16, 83. [Google Scholar] [CrossRef]
- Abbas, T.; Mueller, A.C.; Shibata, E.; Keaton, M.; Rossi, M.; Dutta, A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol. Cell 2013, 49, 1147–1158. [Google Scholar] [CrossRef]
- Abbas, T.; Shibata, E.; Park, J.; Jha, S.; Karnani, N.; Dutta, A. CRL4Cdt2 regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 2010, 40, 9–21. [Google Scholar] [CrossRef]
- Jørgensen, S.; Eskildsen, M.; Fugger, K.; Hansen, L.; Larsen, M.S.Y.; Kousholt, A.N.; Syljuåsen, R.G.; Trelle, M.B.; Jensen, O.N.; Helin, K.; et al. SET8 is degraded via PCNA-coupled CRL4 (CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Boil. 2011, 192, 43–54. [Google Scholar] [CrossRef]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Kamato, D.; Burch, M.L.; Piva, T.J.; Rezaei, H.B.; Rostam, M.A.; Xu, S.; Zheng, W.; Little, P.J.; Osman, N. Transforming growth factor-beta signalling: Role and consequences of Smad linker region phosphorylation. Cell. Signal. 2013, 25, 2017–2024. [Google Scholar] [CrossRef] [PubMed]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Nazish, J.; Wahab, F.; Khan, R.; Jiang, X.; Shi, Q. DDB1 Regulates Sertoli Cell Proliferation and Testis Cord Remodeling by TGFβ Pathway. Genes 2019, 10, 974. https://doi.org/10.3390/genes10120974
Zheng W, Nazish J, Wahab F, Khan R, Jiang X, Shi Q. DDB1 Regulates Sertoli Cell Proliferation and Testis Cord Remodeling by TGFβ Pathway. Genes. 2019; 10(12):974. https://doi.org/10.3390/genes10120974
Chicago/Turabian StyleZheng, Wei, Jabeen Nazish, Fazal Wahab, Ranjha Khan, Xiaohua Jiang, and Qinghua Shi. 2019. "DDB1 Regulates Sertoli Cell Proliferation and Testis Cord Remodeling by TGFβ Pathway" Genes 10, no. 12: 974. https://doi.org/10.3390/genes10120974
APA StyleZheng, W., Nazish, J., Wahab, F., Khan, R., Jiang, X., & Shi, Q. (2019). DDB1 Regulates Sertoli Cell Proliferation and Testis Cord Remodeling by TGFβ Pathway. Genes, 10(12), 974. https://doi.org/10.3390/genes10120974