Nucleolar Expression and Chromosomal Associations in Robertsonian Spermatocytes of Mus musculus domesticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Karyotyping
2.3. Spermatocyte Microspreads
2.4. In-Situ Hybridization (FISH)
2.5. Immunofluorescence (IF)
2.6. Images Analysis
2.7. Statistical Analysis
3. Results
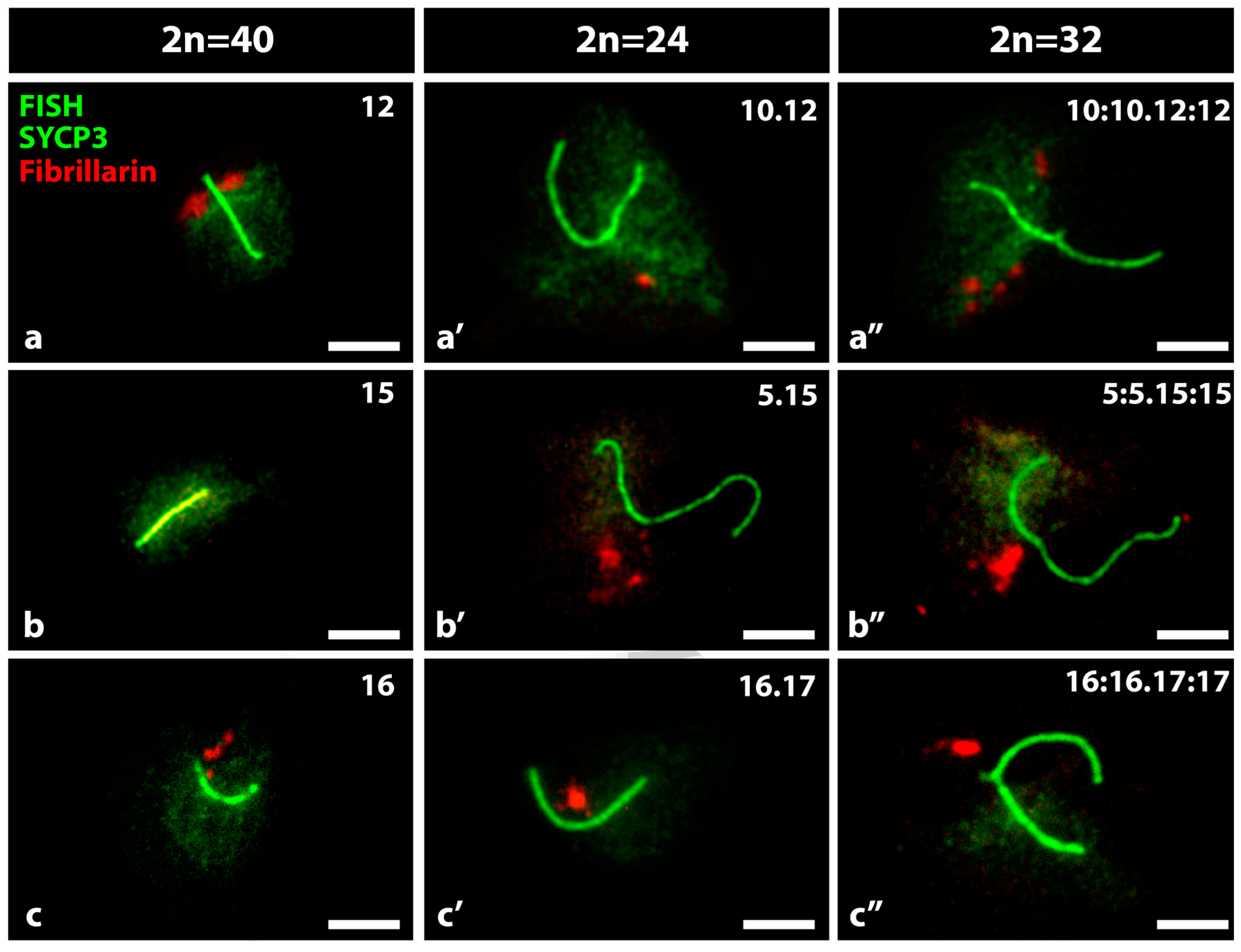
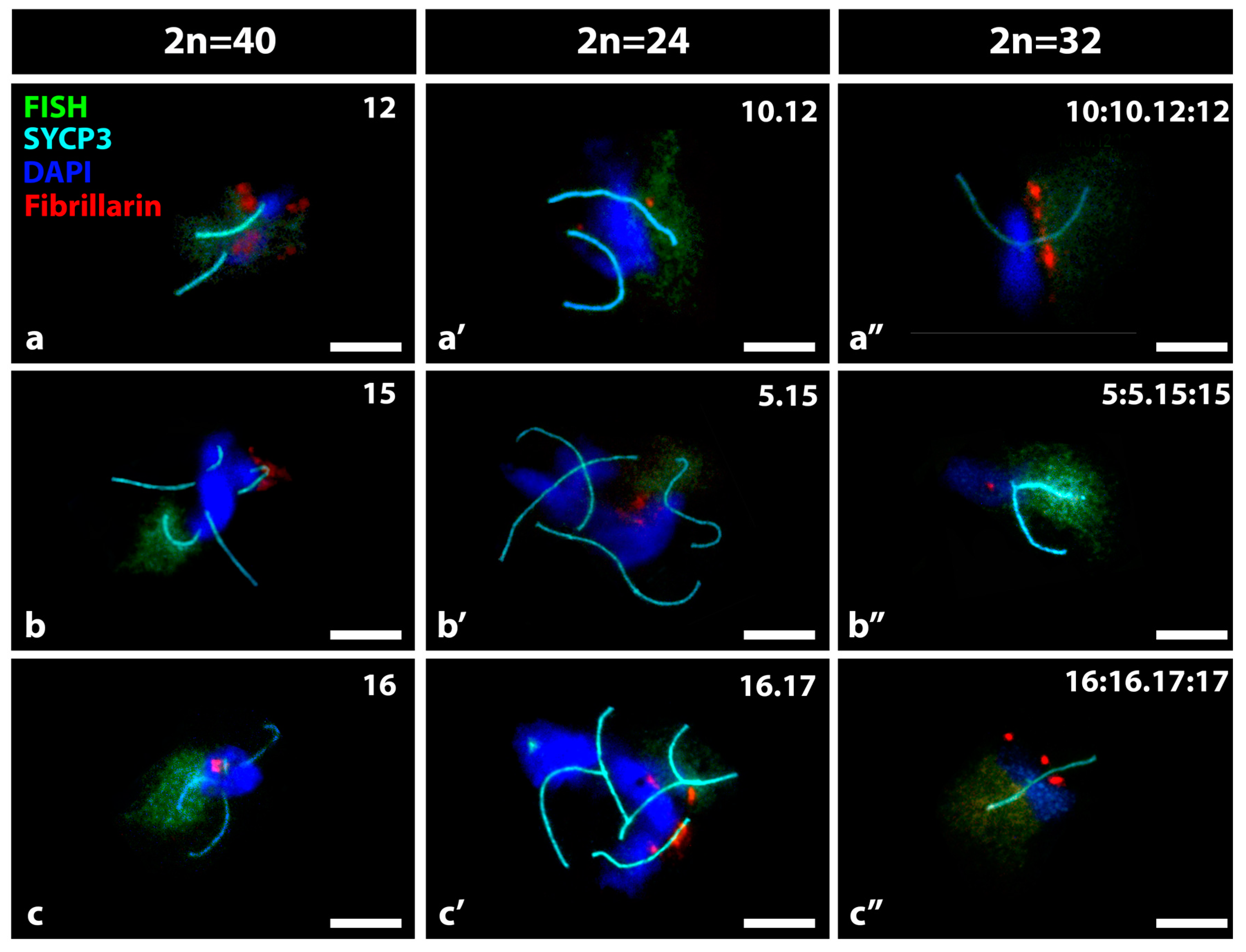
3.1. FISH and IF in the Detection of Nucleolar Expression in Specific Bivalents of Mouse Spermatocytes
3.2. Bivalent Associations from Different Shaped Nucleolar Chromosomes Present in Spermatocytes of Mus m. domesticus 2n = 40, 2n = 24 and 2n = 32
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fakan, S.; Hernandez-Verdun, D. The nucleolus and the nucleolar organizer regions. Biol. Cell 1986, 56, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Scheer, U.; Hock, R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999, 11, 385–390. [Google Scholar] [CrossRef]
- Pederson, T. The nucleolus. Cold Spring Harb Perspect. Biol. 2011, 13, a000638. [Google Scholar] [CrossRef]
- Cazaux, B.; Catalan, J.; Veyrunes, F.; Douzery, E.; Britton-Davidian, J. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae). BMC Evol. Biol. 2011, 11, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Britton-Davidian, J.; Cazaux, B.; Catalan, J. Chromosomal dynamics of nucleolar organizer regions (NORs) in the house mouse: micro-evolutionary insights. Heredity (Edinb). 2012, 108, 68–74. [Google Scholar] [CrossRef]
- Berríos, S.; Fernández-Donoso, R.; Pincheira, J.; Page, J.; Manterola, M.; Cerda, M.C. Number and nuclear localisation of nucleoli in mammalian spermatocytes. Genetica 2004, 121, 219–228. [Google Scholar] [CrossRef]
- López-Fenner, J.; Berríos, S.; Manieu, C.; Fernández-Donoso, R.; Page, J. Bivalent associations in Mus domesticus 2n = 40 spermatocytes. Are they random? Bull. Math. Biol. 2014, 76, 1941–1952. [Google Scholar] [CrossRef]
- Garagna, S.; Redi, C.; Capanna, E.; Andayani, N.; Alfano, R.; Doi, P.; Viale, G. Genome distribution, chromosomal allocation, and organization of the major and minor satellite DNAs in 11 species and subspecies of the genus Mus. Cytogenet. Cell Genet. 1993, 64, 247–255. [Google Scholar] [CrossRef]
- Guenatri, M.; Bailly, D.; Maison, C.; Almouzni, G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004, 166, 493–505. [Google Scholar] [CrossRef]
- Berrios, S. Nuclear architecture of mouse spermatocytes: Chromosome topology, heterochromatin, and nucleolus. Cytogenet. Genome Res. 2017, 151, 61–71. [Google Scholar] [CrossRef]
- Gropp, A.; Winking, H.; Redi, C.; Capanna, E.; Britton-Davidian, J.; Noak, G. Robertsonian karyotype variation in wild house mice from Rhaeto-Lombardia. Cytogenet. Cell Genet. 1982, 34, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Pialek, J.; Hauffe, H.; Searle, J. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 2005, 84, 535–563. [Google Scholar] [CrossRef]
- Elsevier, S.; Ruddle, F. Location of genes coding for 18S and 28S ribosomal RNA within the genome of Mus musculus. Chromosoma 1975, 52, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Garagna, S.; Marziliano, N.; Zuccotti, M.; Searle, J.; Capanna, E.; Redi, C. Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc. Natl. Acad. Sci. USA 2001, 98, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Berríos, S.; Manieu, C.; López-Fenner, J.; Ayarza, E.; Page, J.; González, M.; Manterola, M.; Fernández-Donoso, R. Robertsonian chromosomes and the nuclear architecture of mouse meiotic prophase spermatocytes. Biol. Res. 2014, 47, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, C.; Serebryannyy, L.A.; Feric, M.; Schibler, A.; Meaburn, K.J.; Kubben, N.; Trzaskoma, P.; Shachar, S.; Vidak, S.; Finn, E.H.; et al. Blank spots on the map: some current questions on nuclear organization and genome architecture. Histochem. Cell Biol. 2018, 150, 579–592. [Google Scholar] [CrossRef]
- Peters, A.; Plug, A.; van Vugt, M.; de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom. Res. 1997, 5, 66–68. [Google Scholar] [CrossRef]
- Capanna, E.; Castiglia, R. Chromosomes and speciation in Mus musculus domesticus. Cytogenet. Genome Res. 2004, 105, 375–384. [Google Scholar] [CrossRef]
- Garagna, S.; Page, J.; Fernández-Donoso, R.; Zuccotti, M.; Searle, J. The Robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma 2014, 123, 529–544. [Google Scholar] [CrossRef]
- Grao, P.; Coll, M.; Ponsà, M.; Egozcue, J. Trivalent behavior during prophase I in male mice heterozygous for three Robertsonian translocations: an electron-microscopic study. Cytogenet. Cell Genet. 1989, 152, 105–110. [Google Scholar] [CrossRef]
- Manterola, M.; Page, J.; Vasco, C.; Berríos, S.; Parra, M.; Viera, A.; Rufas, J.; Zuccotti, M.; Garagna, S.; Fernández-Donoso, R. A high incidence of meiotic silencing of asynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet. 2009, 5, e1000625. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T. Higher-order genome organization in human disease. Cold Spring Harb. Perspect. Biol. 2010, 2, a000794. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Cavalli, G. The role of chromosome domains in shaping the functional genome. Cell 2015, 160, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Németh, A.; Grummt, I. Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 2018, 52, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.; Trinkle-Mulcahy, L.; Lamond, A. The nucleolus. J. Cell Sci. 2005, 118, 1335–1337. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H. Imaging of chromosome dynamics in mouse testis tissue by immuno-FISH. In Meiosis; Humana Press: New York, NY, USA, 2017; pp. 231–243. [Google Scholar]
- Farley, K.; Surovtseva, Y.; Merkel, J.; Baserga, S. Determinants of mammalian nucleolar architecture. Chromosoma 2015, 124, 323–331. [Google Scholar] [CrossRef]
- Wolff, D.; Schwartz, S. Characterization of Robertsonian translocations by using fluorescence in situ hybridization. Am. J. Hum. Genet. 1992, 50, 174–181. [Google Scholar]
- Redi, C.; Garagna, S.; Zacharias, H.; Zuccotti, M.; Capanna, E. The other chromatin. Chromosoma 2001, 110, 136–147. [Google Scholar] [CrossRef]
- Peng, J.; Karpen, G. Heterochromatin genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009, 5, e1000435. [Google Scholar] [CrossRef]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Heyting, C. Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol. 1996, 8, 389–396. [Google Scholar] [CrossRef]
- Page, S.; Hawley, R. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004, 20, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Fraune, J.; Schramm, S.; Alsheimer, M.; Benavente, R. The mammalian synaptonmemal complex: protein components, assembly and role in meiotic recombination. Exp. Cell Res. 2012, 318, 1340–1346. [Google Scholar] [CrossRef]
- Zickler, D.; Kleckner, N. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Semin. Cell Dev. Biol. 2016, 54, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Schwarzacher, T.; Mayr, B.; Schweizer, D. Heterochromatin and nucleolus-organizer-region behaviour at male pachytene of Sus scrofa domestica. Chromosoma 1984, 91, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.; Tantravahi, R.; Dev, V.; Miller, O. Frequency of satellite association of human chromosomes is correlated with amount of Ag-staining of the nucleolus organizer region. Am. J. Hum. Genet. 1977, 29, 490–502. [Google Scholar]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Berrios, S.; Manterola, M.; Prieto, Z.; López-Fenner, J.; Fernández-Donoso, R. Model of chromosome associations in Mus domesticus spermatocytes. Biol. Res. 2010, 43, 275–285. [Google Scholar] [CrossRef]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Moncada, F.; Tapia, D.; Zuñiga, N.; Ayarza, E.; López-Fenner, J.; Redi, C.A.; Berríos, S. Nucleolar Expression and Chromosomal Associations in Robertsonian Spermatocytes of Mus musculus domesticus. Genes 2019, 10, 120. https://doi.org/10.3390/genes10020120
López-Moncada F, Tapia D, Zuñiga N, Ayarza E, López-Fenner J, Redi CA, Berríos S. Nucleolar Expression and Chromosomal Associations in Robertsonian Spermatocytes of Mus musculus domesticus. Genes. 2019; 10(2):120. https://doi.org/10.3390/genes10020120
Chicago/Turabian StyleLópez-Moncada, Fernanda, Daniel Tapia, Nolberto Zuñiga, Eliana Ayarza, Julio López-Fenner, Carlo Alberto Redi, and Soledad Berríos. 2019. "Nucleolar Expression and Chromosomal Associations in Robertsonian Spermatocytes of Mus musculus domesticus" Genes 10, no. 2: 120. https://doi.org/10.3390/genes10020120




