Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Marker Choice and Primer Design
2.2. Samples
2.3. DNA Extraction and Selection of the Optimal Extraction Method
2.4. DNA Library Preparation
2.5. Data Analysis
2.6. Test of the Primers on Individual Components
2.7. Data availability Statement
3. Results and Discussion
3.1. Comparison of the DNA Extraction Methods
3.2. Primer Design and Amplification of nrITS1
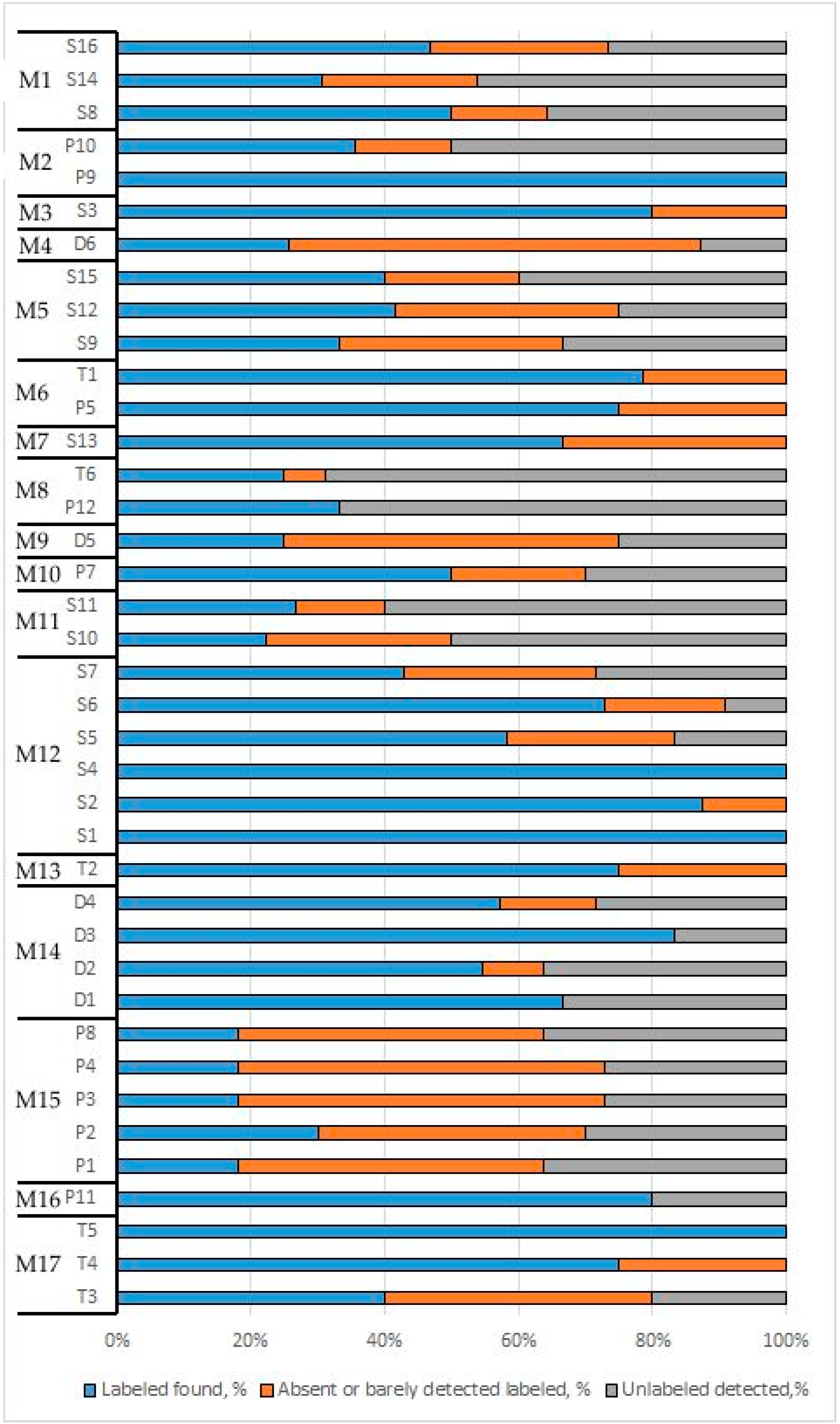
3.3. Composition Analysis of Food Products
3.4. Economical Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Egan, B.; Hodgkins, C.; Shepherd, R.; Timotijevic, L.; Raats, M. An overview of consumer attitudes and beliefs about plant food supplements. Food Funct. 2011, 2, 747. [Google Scholar] [CrossRef] [PubMed]
- Mintel Press Team Taste is the Top Reason US Consumers Eat Plant-Based Proteins. Available online: http://www.mintel.com/press-centre/food-and-drink/taste-is-the-top-reason-us-consumers-eat-plant-based-proteins (accessed on 6 February 2019).
- Siegner, C. Consumers Reveal Why They Buy Plant-Based Dairy Alternatives. Available online: https://www.fooddive.com/news/consumers-reveal-why-they-buy-plant-based-dairy-alternatives/516702/ (accessed on 6 February 2019).
- Cupp, M.J. Toxicology and Clinical Pharmacology of Herbal Products; Forensic Science and Medicine; Humana Press: Totowa, NJ, USA, 2000; ISBN 978-0-89603-791-5. [Google Scholar]
- Di Lorenzo, C.; Ceschi, A.; Kupferschmidt, H.; Lüde, S.; De Souza Nascimento, E.; Dos Santos, A.; Colombo, F.; Frigerio, G.; Nørby, K.; Plumb, J.; et al. Adverse effects of plant food supplements and botanical preparations: A systematic review with critical evaluation of causality. Br. J. Clin. Pharmacol. 2015, 79, 578–592. [Google Scholar] [CrossRef]
- de Boer, H.J.; Ghorbani, A.; Manzanilla, V.; Raclariu, A.-C.; Kreziou, A.; Ounjai, S.; Osathanunkul, M.; Gravendeel, B. DNA metabarcoding of orchid-derived products reveals widespread illegal orchid trade. Proc. R. Soc. B 2017, 284, 20171182. [Google Scholar] [CrossRef] [PubMed]
- Newmaster, S.G.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Gamble, C.C.; Kirpekar, R.; Young, G.; Ahmed, S.; Little, D.P. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci. Rep. 2011, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Vassou, S.L.; Kusuma, G.; Parani, M. DNA barcoding for species identification from dried and powdered plant parts: A case study with authentication of the raw drug market samples of Sida cordifolia. Gene 2015, 559, 86–93. [Google Scholar] [CrossRef]
- Speranskaya, A.S.; Khafizov, K.; Ayginin, A.A.; Krinitsina, A.A.; Omelchenko, D.O.; Nilova, M.V.; Severova, E.E.; Samokhina, E.N.; Shipulin, G.A.; Logacheva, M.D. Comparative analysis of Illumina and Ion Torrent high-throughput sequencing platforms for identification of plant components in herbal teas. Food Control 2018, 93, 315–324. [Google Scholar] [CrossRef]
- Desalle, R.; Birstein, V.J. PCR identification of black caviar. Nature 1996, 381, 197–198. [Google Scholar] [CrossRef]
- Willette, D.A.; Simmonds, S.E.; Cheng, S.H.; Esteves, S.; Kane, T.L.; Nuetzel, H.; Pilaud, N.; Rachmawati, R.; Barber, P.H. Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. Conserv. Biol. 2017, 31, 1076–1085. [Google Scholar] [CrossRef]
- Kane, D.E.; Hellberg, R.S. Identification of species in ground meat products sold on the U.S. commercial market using DNA-based methods. Food Control 2016, 59, 158–163. [Google Scholar] [CrossRef]
- Eurofins Eurofins Meat Species Testing. Available online: https://www.eurofins.co.uk/food-testing/food-authenticity/meat-species-testing/ (accessed on 18 January 2019).
- U. S. Fish and Wildlife Service Caviar Company, Corporate Officers Sentenced for Illegal Trade, Fraud Scheme; Landmark Fine Assessed. Available online: https://www.fws.gov/news/ShowNews.cfm?ID=94F51E9E-CA53-4419-A23D7870E0F7CA01 (accessed on 18 January 2019).
- Doukakis, P.; Pikitch, E.K.; Rothschild, A.; DeSalle, R.; Amato, G.; Kolokotronis, S.-O. Testing the Effectiveness of an International Conservation Agreement: Marketplace Forensics and CITES Caviar Trade Regulation. PLoS ONE 2012, 7, e40907. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, S.; Blaschitz, M.; Weinmaier, T.; Pechatschek, J.; Cichna-Markl, M.; Indra, A.; Hufnagl, P.; Hochegger, R. Development of a DNA metabarcoding method for the identification of fifteen mammalian and six poultry species in food. Food Chem. 2019, 272, 354–361. [Google Scholar] [CrossRef] [PubMed]
- de Boer, H.J.; Ichim, M.C.; Newmaster, S.G. DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf. 2015, 38, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.W.J.; Hebert, P.D.N. Rapid identification of the botanical and entomological sources of honey using DNA metabarcoding. Food Chem. 2017, 214, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Sánchez-Hernández, C.; Gaytán-Oyarzún, J.C. Two mini-preparation protocols to DNA extraction from plants with high polysaccharide and secondary metabolites. Afr. J. Biotechnol. 2006, 5, 1864–1867. [Google Scholar]
- Staats, M.; Cuenca, A.; Richardson, J.E.; Ginkel, R.V.; Petersen, G.; Seberg, O.; Bakker, F.T. DNA Damage in Plant Herbarium Tissue. PLoS ONE 2011, 6, e28448. [Google Scholar] [CrossRef]
- Gryson, N. Effect of food processing on plant DNA degradation and PCR-based GMO analysis: A review. Anal. Bioanal. Chem. 2010, 396, 2003–2022. [Google Scholar] [CrossRef]
- Bernardo, G.D.; Gaudio, S.D.; Galderisi, U.; Cascino, A.; Cipollaro, M. Comparative Evaluation of Different DNA Extraction Procedures from Food Samples. Biotechnol. Prog. 2007, 23, 297–301. [Google Scholar] [CrossRef]
- Costa, J.; Melo, V.S.; Santos, C.G.; Oliveira, M.B.P.P.; Mafra, I. Tracing tree nut allergens in chocolate: A comparison of DNA extraction protocols. Food Chem. 2015, 187, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Pafundo, S.; Gullì, M.; Marmiroli, N. Comparison of DNA Extraction Methods and Development of Duplex PCR and Real-Time PCR To Detect Tomato, Carrot, and Celery in Food. J. Agric. Food Chem. 2011, 59, 10414–10424. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.D.; Forte, V.; Guastadisegni, M.C.; Martino, C.; Schena, F.P.; Tantillo, G. A comparison of DNA extraction methods for food analysis. Food Control 2007, 18, 76–80. [Google Scholar] [CrossRef]
- Smith, D.S.; Maxwell, P.W.; De Boer, S.H. Comparison of several methods for the extraction of DNA from potatoes and potato-derived products. J. Agric. Food Chem. 2005, 53, 9848–9859. [Google Scholar] [CrossRef] [PubMed]
- Takabatake, R.; Noritake, H.; Noguchi, A.; Nakamura, K.; Kondo, K.; Akiyama, H.; Teshima, R.; Mano, J.; Kitta, K. Comparison of DNA extraction methods for sweet corn and processed sweet corns. Shokuhin Eiseigaku Zasshi J. Food Hyg. Soc. Jpn. 2013, 54, 309–315. [Google Scholar] [CrossRef]
- Wang, X.-C.; Liu, C.; Huang, L.; Bengtsson-Palme, J.; Chen, H.; Zhang, J.-H.; Cai, D.; Li, J.-Q. ITS1: A DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Resour. 2015, 15, 573–586. [Google Scholar] [CrossRef]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef]
- Moorhouse-Gann, R.J.; Dunn, J.C.; de Vere, N.; Goder, M.; Cole, N.; Hipperson, H.; Symondson, W.O.C. New universal ITS2 primers for high-resolution herbivory analyses using DNA metabarcoding in both tropical and temperate zones. Sci. Rep. 2018, 8, 8542. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- China Plant BOL Group; Li, D.-Z.; Gao, L.-M.; Li, H.-T.; Wang, H.; Ge, X.-J.; Liu, J.-Q.; Chen, Z.-D.; Zhou, S.-L.; Chen, S.-L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar]
- National Plant Data Team, Greensboro, NC 27401-4901 USA. USDA; NRCS; The PLANTS Database; 2019. Available online: http://plants.usda.gov (accessed on 6 February 2019).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Tissot, C.; Schmidt, E.; Pechtl, I.; Gassmann, M. The DNA Integrity Number: A novel approach for objective integrity classification of genomic DNA samples. Chromosome Res. 2015, 23, S143. [Google Scholar]
- Krinitsina, A.A.; Sizova, T.V.; Zaika, M.A.; Speranskaya, A.S.; Sukhorukov, A.P. A rapid and cost-effective method for DNA extraction from archival herbarium specimens. Biochem. Mosc. 2015, 80, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Seregin, A. Moscow University Herbarium (MW). Available online: https://plant.depo.msu.ru/?d=P (accessed on 6 February 2019).
- Seregin, A.P. Making the Russian Flora Visible: Fast Digitisation of the Moscow University Herbarium (MW) in 2015. TAXON 2016, 65, 205–207. [Google Scholar] [CrossRef]
- ID 486584-BioProject-NCBI. Available online: https://www.ncbi.nlm.nih.gov/bioproject/486584 (accessed on 18 January 2019).
- Shmakov, A.I.; Kutsev, M.G.; Sinitsyna, T.A.; Uvarova, O.V.; Smirnov, S.V.; Kondo, K. Taxonomic rank of Tanacetum boreale Fisch. ex DC. (Asteraceae). Chromosome Bot. 2017, 11, 72–76. [Google Scholar] [CrossRef]
- Mudrik, E.A.; Polyakova, T.A.; Shatokhina, A.V.; Bondarenko, G.N.; Politov, D.V. Spatial distribution of intron 2 of nad1 gene haplotypes in populations of Norway and Siberian spruce (Picea abies–P. obovata) species complex. Russ. J. Genet. 2015, 51, 957–965. [Google Scholar] [CrossRef]
- Gudkova, P.; Bayahmetov, E. Optimization of Polymerase Chain Reaction for Inter Simple Sequence Repeat Technique for Four Species of Plants. Key Eng. Mater. 2016, 683. [Google Scholar] [CrossRef]
- Sukhorukov, A.P.; Nilova, M.V.; Erst, A.S.; Kushunina, M.; Baider, C.; Verloove, F.; Salas-Pascual, M.; Belyaeva, I.V.; Krinitsina, A.A.; Bruyns, P.V. Diagnostics, taxonomy, nomenclature and distribution of perennial Sesuvium (Aizoaceae) in Africa. PhytoKeys 2018, 45. [Google Scholar] [CrossRef] [PubMed]
- Llongueras, J.P.; Nair, S.; Salas-Leiva, D.; Schwarzbach, A.E. Comparing DNA Extraction Methods for Analysis of Botanical Materials Found in Anti-Diabetic Supplements. Mol. Biotechnol. 2013, 53, 249–256. [Google Scholar] [CrossRef]
- Rathnayake, A.S.; Allué, J.; Llugany, M.; Puig-Pujol, A.; Hirimburegama, K.; Poschenrieder, C. High quality DNA obtained from a single seed of Vitis vinifera L. using rapid DNA extraction method. Am. J. Plant Sci. 2014, 5, 2023. [Google Scholar] [CrossRef]
- Verbylaitė, R.; Beišys, P.; Rimas, V.; Kuusienė, S. Comparison of ten DNA extraction protocols from wood of European aspen (Populus tremula L.). Balt. For. 2010, 16, 35–42. [Google Scholar]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Su, X.; Chen, X.; Zhao, H.; Bo, C.; Xu, J.; Bai, H.; Ning, K. Biological ingredient analysis of traditional Chinese medicine preparation based on high-throughput sequencing: The story for Liuwei Dihuang Wan. Sci. Rep. 2014, 4, 5147. [Google Scholar] [CrossRef] [PubMed]
- Raclariu, A.C.; Paltinean, R.; Vlase, L.; Labarre, A.; Manzanilla, V.; Ichim, M.C.; Crisan, G.; Brysting, A.K.; Boer, H. de Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 2017, 7, 1291. [Google Scholar] [CrossRef] [PubMed]
- Meira, M.; da Silva, E.P.; David, J.M.; David, J.P. Review of the genus Ipomoea: Traditional uses, chemistry and biological activities. Rev. Bras. Farmacogn. 2012, 22, 682–713. [Google Scholar] [CrossRef]
- Sõukand, R.; Quave, C.L.; Pieroni, A.; Pardo-de-Santayana, M.; Tardío, J.; Kalle, R.; Łuczaj, Ł.; Svanberg, I.; Kolosova, V.; Aceituno-Mata, L.; et al. Plants used for making recreational tea in Europe: A review based on specific research sites. J. Ethnobiol. Ethnomed. 2013, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Pillitteri, J.L.; Shiffman, S.; Rohay, J.M.; Harkins, A.M.; Burton, S.L.; Wadden, T.A. Use of Dietary Supplements for Weight Loss in the United States: Results of a National Survey. Obesity 2008, 16, 790–796. [Google Scholar] [CrossRef]
- Steinhoff, B.; Programme, T.M. Regulatory Situation of Herbal Medicines: A Worldwide Review; World Health Organization: Geneva, Switzerland, 1998; ISBN 978-0-11-987569-0. [Google Scholar]
- Everstine, K.; Spink, J.; Kennedy, S. Economically Motivated Adulteration (EMA) of Food: Common Characteristics of EMA Incidents. J. Food Prot. 2013, 76, 723–735. [Google Scholar] [CrossRef]



| Genus Labelled by Manufacturer | Illumina not Found | Ion Torrent not Found | Illumina Found Below the Threshold | Ion Torrent Found below Threshold | Median GC-Content, % | GC-Content IQR |
|---|---|---|---|---|---|---|
| Allium | 8 of 10 | 8 of 10 | 2 of 10 | 1 of 10 | 43.3 | 5.2 |
| Piper | 3 of 9 | 5 of 9 | 5 of 9 | 1 of 9 | 54.9 | 6.8 |
| Vaccinium | 3 of 5 | 3 of 5 | ND | ND | 57.1 | 2.1 |
| Equisetum | 3 of 3 | 3 of 3 | ND | ND | 67.1 | 1.8 |
| Rosa | 2 of 7 | 3 of 7 | 2 of 7 | 1 of 7 | 57.6 | 1.6 |
| Matricaria | 2 of 5 | 2 of 5 | ND | ND | 46.0 | 1.0 |
| Berberis | 2 of 3 | 1 of 3 | ND | ND | 45.9 | 1.1 |
| Orthosiphon | 2 of 2 | 2 of 2 | ND | ND | 65.9 | 0.9 |
| Capsicum | 1 of 9 | 8 of 9 | 5 of 9 | 1 of 9 | 52.2 | 13.9 |
| Curcuma | ND | 5 of 5 | 5 of 5 | ND | 53.0 | 1.8 |
| Detected Unlabelled Plants | Illumina | Ion Torrent | Note |
|---|---|---|---|
| Elymus | 7 of 39 | 6 of 39 | field weed |
| Triticum | 6 of 39 | 7 of 39 | food plant |
| Brassica | 5 of 39 | 5 of 39 | food plant |
| Secale | 5 of 39 | 5 of 39 | food plant |
| Convolvulus | 6 of 39 | 3 of 39 | field weed |
| Coriandrum | 4 of 39 | 4 of 39 | food plant |
| Calystegia | 4 of 39 | 4 of 39 | field weed |
| Ambrosia | 4 of 39 | 3 of 39 | invasive weed |
| Panicum | 4 of 39 | 3 of 39 | food plant |
| Helosciadium | 3 of 39 | 4 of 39 | food plant |
| Medicago | 4 of 39 | 2 of 39 | field weed/forage plant |
| Zea | 3 of 39 | 3 of 39 | food plant |
| Ocimum | 3 of 39 | 2 of 39 | food plant |
| Rorippa | 3 of 39 | 2 of 39 | field weed |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omelchenko, D.O.; Speranskaya, A.S.; Ayginin, A.A.; Khafizov, K.; Krinitsina, A.A.; Fedotova, A.V.; Pozdyshev, D.V.; Shtratnikova, V.Y.; Kupriyanova, E.V.; Shipulin, G.A.; et al. Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products. Genes 2019, 10, 122. https://doi.org/10.3390/genes10020122
Omelchenko DO, Speranskaya AS, Ayginin AA, Khafizov K, Krinitsina AA, Fedotova AV, Pozdyshev DV, Shtratnikova VY, Kupriyanova EV, Shipulin GA, et al. Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products. Genes. 2019; 10(2):122. https://doi.org/10.3390/genes10020122
Chicago/Turabian StyleOmelchenko, Denis O., Anna S. Speranskaya, Andrey A. Ayginin, Kamil Khafizov, Anastasia A. Krinitsina, Anna V. Fedotova, Denis V. Pozdyshev, Viktoria Y. Shtratnikova, Evgenia V. Kupriyanova, German A. Shipulin, and et al. 2019. "Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products" Genes 10, no. 2: 122. https://doi.org/10.3390/genes10020122
APA StyleOmelchenko, D. O., Speranskaya, A. S., Ayginin, A. A., Khafizov, K., Krinitsina, A. A., Fedotova, A. V., Pozdyshev, D. V., Shtratnikova, V. Y., Kupriyanova, E. V., Shipulin, G. A., & Logacheva, M. D. (2019). Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products. Genes, 10(2), 122. https://doi.org/10.3390/genes10020122






