Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Harvest
2.2. Library Preparation and RNA Sequencing
2.3. Transcript Quantification, Differential Expression Analysis, and Gene Annotation
2.4. Quantitative Real-Time Polymerase Chain Reaction
2.5. Data Deposition
3. Results
3.1. Sequencing and Statistical Evaluation of the Brassica rapa Transcriptome
3.2. Identification of Differentially Expressed Genes in Pairwise Sample Comparisons
3.3. Conserved Phasing of Diurnally Expressed Genes in B. rapa
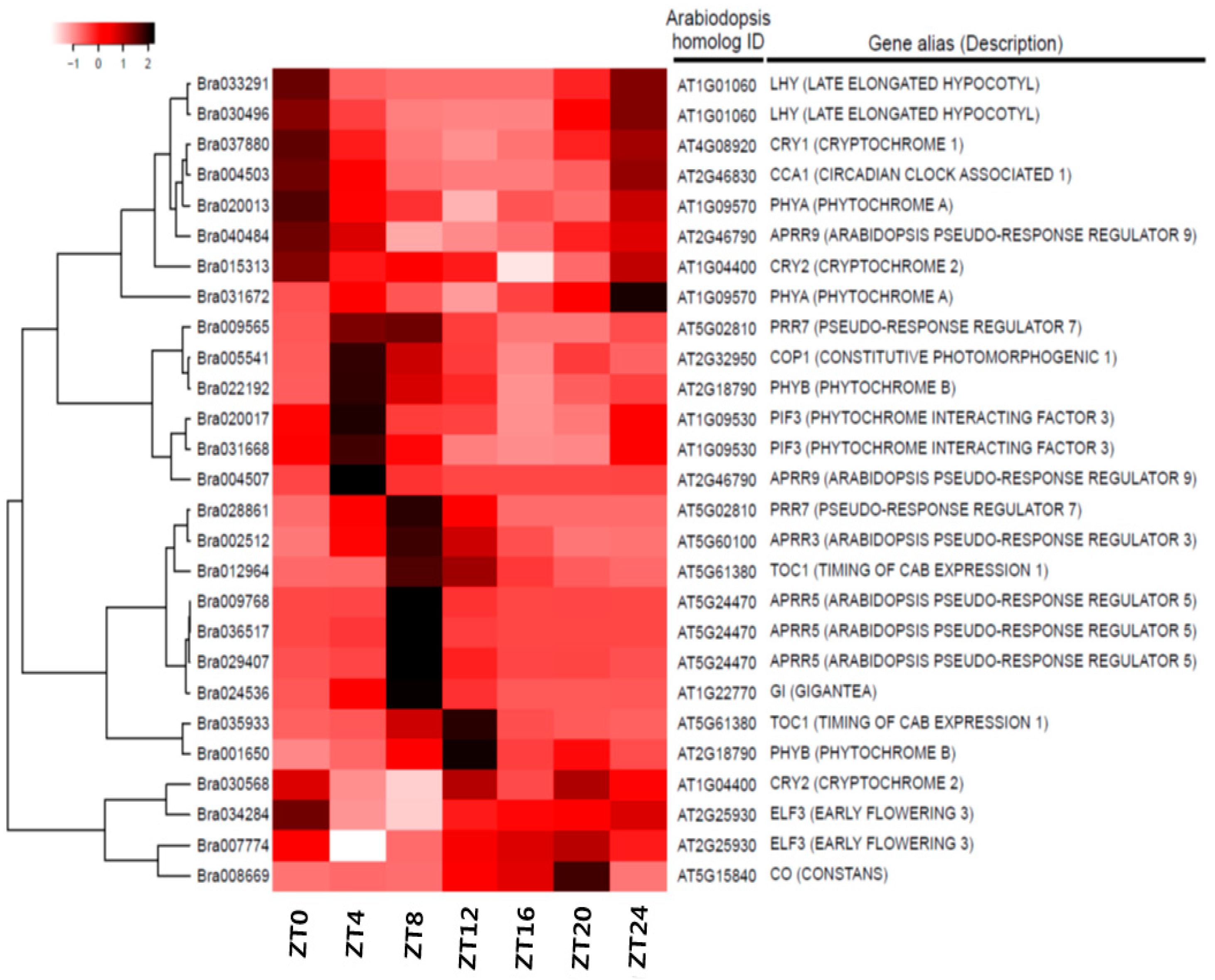
3.4. Expression Anaysis of Clock-Related Gene Paralogs in Chinese Cabbage
3.5. Differentially Expression of Glucosinolate-Related Gene Paralogs in Chinese Cabbage
4. Discussion
4.1. Circadian-Regulated Genes in the Brassica Genome
4.2. Circadian Genes Conserved in Brassica after Genome Duplication Events
4.3. Expression Patterns of Paralogous Gene Copies
4.4. Regulation of Circadian-Mediated Biological Processes in Crops
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hangarter, R.P. Gravity, light and plant form. Plant Cell Environ. 1997, 20, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.E.; Millar, A.J. The circadian clock. A plant’s best friend in a spinning world. Plant Physiol. 2003, 132, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Yakir, E.; Hilman, D.; Harir, Y.; Green, R.M. Regulation of output from the plant circadian clock. FEBS J. 2007, 274, 335–345. [Google Scholar] [CrossRef]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Fenske, M.P.; Imaizumi, T. Circadian Rhythms in Floral Scent Emission. Front. Plant Sci. 2016, 7, 462. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Shin, J.; Davis, S.J. Abiotic stress and the plant circadian clock. Plant Signal. Behav. 2011, 6, 223–231. [Google Scholar] [CrossRef]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Lin, C. Blue light receptors and signal transduction. Plant Cell 2002, 14, S207–S225. [Google Scholar] [CrossRef]
- Kircher, S.; Kozma-Bognar, L.; Kim, L.; Adam, E.; Harter, K.; Schafer, E.; Nagy, F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 1999, 11, 1445–1456. [Google Scholar] [CrossRef]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Bahler, J. RNA-seq: From technology to biology. Cell Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.E.; Grant, G.R.; Paquin, C.; Qian, J.; Nitabach, M.N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012, 22, 1266–1281. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.; Aoki, K.; Nagano, A.J.; Honjo, M.N.; Fukuda, H. Circadian oscillation of the lettuce transcriptome under constant light and light–dark conditions. Front. Plant Sci. 2016, 7, 1114. [Google Scholar] [CrossRef] [PubMed]
- Facella, P.; Lopez, L.; Carbone, F.; Galbraith, D.W.; Giuliano, G.; Perrotta, G. Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE 2008, 3, e2798. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Breton, G.; Priest, H.D.; Dharmawardhana, P.; Jaiswal, P.; Fox, S.E.; Michael, T.P.; Chory, J.; Kay, S.A.; Mockler, T.C. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 2011, 6, e16907. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Comes a time. Curr. Opin. Plant Biol. 2008, 11, 514–520. [Google Scholar] [CrossRef]
- Farre, E.M.; Weise, S.E. The interactions between the circadian clock and primary metabolism. Curr. Opin. Plant Biol. 2012, 15, 293–300. [Google Scholar] [CrossRef]
- Farre, E.M. The regulation of plant growth by the circadian clock. Plant Biol. (Stuttg.) 2012, 14, 401–410. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, H.S.; Choi, S.H.; Jang, J.Y.; Jeong, M.J.; Lee, S.I. The importance of the circadian clock in regulating plant metabolism. Int. J. Mol. Sci. 2017, 18, 2680. [Google Scholar] [CrossRef]
- Michael, T.P.; Mockler, T.C.; Breton, G.; McEntee, C.; Byer, A.; Trout, J.D.; Hazen, S.P.; Shen, R.; Priest, H.D.; Sullivan, C.M.; et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008, 4, e14. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, T.L.; Field, C.B. Circadian rhythms in photosynthesis: Oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol. 1991, 96, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Rikin, A.; Dillwith, J.W.; Bergman, D.K. Correlation between the circadian rhythm of resistance to extreme temperatures and changes in fatty acid composition in cotton seedlings. Plant Physiol. 1993, 101, 31–36. [Google Scholar] [CrossRef]
- Hudson, K.A. The circadian clock-controlled transcriptome of developing soybean seeds. Plant Genome-US 2010, 3, 3–13. [Google Scholar] [CrossRef]
- Kim, J.A.; Jung, H.E.; Hong, J.K.; Hermand, V.; Robertson McClung, C.; Lee, Y.H.; Kim, J.Y.; Lee, S.I.; Jeong, M.J.; Kim, J.; et al. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA Confers Robust Diurnal Rhythms on the Global Transcriptome of Rice in the Field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef]
- Xie, Q.G.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C.; et al. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, S.P.; Lan, T.H.; Feldmann, K.A.; Paterson, A.H. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 1994, 138, 499–510. [Google Scholar]
- Grant, D.; Cregan, P.; Shoemaker, R.C. Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 4168–4173. [Google Scholar] [CrossRef]
- Bowers, J.E.; Abbey, C.; Anderson, S.; Chang, C.; Draye, X.; Hoppe, A.H.; Jessup, R.; Lemke, C.; Lennington, J.; Li, Z.K.; et al. A high-density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 2003, 165, 367–386. [Google Scholar]
- Gebhardt, C.; Walkemeier, B.; Henselewski, H.; Barakat, A.; Delseny, M.; Stuber, K. Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J. 2003, 34, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Salathia, N.; Lynn, J.R.; Millar, A.J.; King, G.J. Detection and resolution of genetic loci affecting circadian period in Brassica oleracea. Theor. Appl. Genet. 2007, 114, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Wolfe, K.H. Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 2008, 9, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Hou, S.B.; Feng, Y.; Yu, Q.Y.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Yang, T.J.; Seol, Y.J.; Jin, M.; Kim, J.A.; Lim, M.H.; Kim, J.S.; Baek, S.; Choi, B.S.; et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.H.; Kwon, S.J.; Seol, Y.J.; Kim, J.A.; Jin, M.; Kim, J.S.; Lim, M.H.; Lee, S.I.; Hong, J.K.; Park, T.H.; et al. Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 2010, 11, R94. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Saito, S.; Saito, C.T.; Uemura, M. Phylogenetic footprint of the plant clock system in angiosperms: Evolutionary processes of pseudo-response regulators. BMC Evol. Biol. 2010, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Kim, J.S.; Hong, J.K.; Lee, Y.H.; Choi, B.S.; Seol, Y.J.; Jeon, C.H. Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol. Genet. Genomics 2012, 287, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Wu, J.; Cheng, F.; Cressman, L.G.; Wang, X.W.; McClung, C.R. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 2012, 24, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.Y.; Lee, S.; Seo, P.J.; Yang, M.S.; Park, C.M. Identification and molecular characterization of a Brachypodium distachyon GIGANTEA gene: Functional conservation in monocot and dicot plants. Plant Mol. Biol. 2010, 72, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Chung, T.Y.; King, G.J.; Jin, M.; Yang, T.J.; Jin, Y.M.; Kim, H.I.; Park, B.S. A sequence-tagged linkage map of Brassica rapa. Genetics 2006, 174, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Campo, C. Biology of Brassica Coenospecies, 1st ed.; Elsevier: Amsterdam, The Netherlands; New York, NY, USA, 1999. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 2013, 14, 689. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11–17. [Google Scholar]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008, 9, 321–332. [Google Scholar] [CrossRef]
- Hughes, M.E.; Hogenesch, J.B.; Kornacker, K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 2010, 25, 372–380. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Chen, Y.H.; Wang, Z.Y.; Chen, Z.L.; Gu, H.Y.; Qu, L.J. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 2007, 51, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Fernandez, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef]
- Takase, T.; Nishiyama, Y.; Tanihigashi, H.; Ogura, Y.; Miyazaki, Y.; Yamada, Y.; Kiyosue, T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011, 67, 608–621. [Google Scholar] [CrossRef]
- Schuster, J.; Knill, T.; Reichelt, M.; Gershenzon, J.; Binder, S. BRANCHED-CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 2006, 18, 2664–2679. [Google Scholar] [CrossRef]
- Zang, Y.X.; Kim, H.U.; Kim, J.A.; Lim, M.H.; Jin, M.; Lee, S.C.; Kwon, S.J.; Lee, S.I.; Hong, J.K.; Park, T.H.; et al. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2009, 276, 3559–3574. [Google Scholar] [CrossRef]
- Mockler, T.C.; Michael, T.P.; Priest, H.D.; Shen, R.; Sullivan, C.M.; Givan, S.A.; McEntee, C.; Kay, S.A.; Chory, J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Blasing, O.E.; Gibon, Y.; Gunther, M.; Hohne, M.; Morcuende, R.; Osuna, D.; Thimm, O.; Usadel, B.; Scheible, W.R.; Stitt, M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 2005, 17, 3257–3281. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; He, K.; Liu, D.; Bai, S.N.; Gu, X.C.; Wei, L.P.; Luo, J.C. DATF: A database of Arabidopsis transcription factors. Bioinformatics 2005, 21, 2568–2569. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Seki, M.; Sakurai, T.; Satou, M.; Akiyama, K.; Toyoda, T.; Konagaya, A.; Shinozaki, K. RARTF: Database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Davuluri, R.V.; Sun, H.; Palaniswamy, S.K.; Matthews, N.; Molina, C.; Kurtz, M.; Grotewold, E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform. 2003, 4, 25. [Google Scholar] [CrossRef]
- Riano-Pachon, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef]
- Carre, I.A.; Kim, J.Y. MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 2002, 53, 1551–1557. [Google Scholar] [CrossRef]
- Green, R.M.; Tobin, E.M. The role of CCA1 and LHY in the plant circadian clock. Dev. Cell 2002, 2, 516–518. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef]
- Jiao, Y.L.; Yang, H.J.; Ma, L.G.; Sun, N.; Yu, H.Y.; Liu, T.; Gao, Y.; Gu, H.Y.; Chen, Z.L.; Wada, M.; et al. A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol. 2003, 133, 1480–1493. [Google Scholar] [CrossRef]
- Hofmann, N.R. Evolution of the circadian clock in a whole-genome context. Plant Cell 2012, 24, 2239. [Google Scholar] [CrossRef]
- McClung, C.R. The genetics of plant clocks. Adv. Genet. 2011, 74, 105–139. [Google Scholar] [PubMed]
- Kuno, N.; Moller, S.G.; Shinomura, T.; Xu, X.M.; Chua, N.H.; Furuya, M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 2003, 15, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.F.; Andersson, C.R.; Zhao, Y.D.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Satbhai, S.B.; Yamashino, T.; Mizuno, T.; Aoki, S. Heterologous expression and functional characterization of a Physcomitrella pseudo response regulator homolog, PpPRR2, in Arabidopsis. Biosci. Biotechnol. Biochem. 2011, 75, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Para, A.; Farre, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 2000, 101, 319–329. [Google Scholar] [CrossRef]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-Box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Lasswell, J.; Rogg, L.E.; Cohen, M.A.; Bartel, B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 2000, 101, 331–340. [Google Scholar] [CrossRef]
- McClung, C.R. A modern circadian clock in the common angiosperm ancestor of monocots and eudicots. BMC Biol. 2010, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Baumlein, H.; Mock, H.P.; Himmelbach, A.; Schweizer, P. The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Markov, G.V.; Paris, M.; Bertrand, S.; Laudet, V. The evolution of the ligand/receptor couple: A long road from comparative endocrinology to comparative genomics. Mol. Cell Endocrinol. 2008, 293, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Huang, Y.; Liu, C.; Ruan, Y.; Shen, W.H. Functional conservation and divergence of J-domain-containing ZUO1/ZRF orthologs throughout evolution. Planta 2014, 239, 1159–1173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fan, T.; Song, J.; Sun, W.; Xia, K.; Liao, J.; Zhang, M. Functional conservation and divergence of four ginger AP1/AGL9 MADS-box genes revealed by analysis of their expression and protein-protein interaction, and ectopic expression of AhFUL gene in Arabidopsis. PLoS ONE 2014, 9, e114134. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Ewers, B.E.; Williams, D.G.; Xie, Q.; Lou, P.; Xu, X.; McClung, C.R.; Weinig, C. The genetic architecture of ecophysiological and circadian traits in Brassica rapa. Genetics 2011, 189, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, Y.; Yeom, M.; Kim, J.H.; Nam, H.G. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 2008, 20, 307–319. [Google Scholar] [CrossRef]
- Huseby, S.; Koprivova, A.; Lee, B.R.; Saha, S.; Mithen, R.; Wold, A.B.; Bengtsson, G.B.; Kopriva, S. Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis. J. Exp. Bot. 2013, 64, 1039–1048. [Google Scholar] [CrossRef]
- Kerwin, R.E.; Jimenez-Gomez, J.M.; Fulop, D.; Harmer, S.L.; Maloof, J.N.; Kliebenstein, D.J. Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 2011, 23, 471–485. [Google Scholar] [CrossRef] [PubMed]






| Index | Sample Name | Num. of Raw Reads | Num. of Cleaned Reads | Mean Length of Cleaned Reads (bp) | Num. of Mapped Reads to Brassica rapa Transcripts | Percent of Mapped Reads |
|---|---|---|---|---|---|---|
| 1 | ZT0_rep1 | 102,435,614 | 102,408,172 | 98.25 | 73,819,707 | 72.08% |
| 2 | ZT0_rep2 | 96,521,352 | 96,496,334 | 98.28 | 69,622,768 | 72.15% |
| 3 | ZT4_rep1 | 87,345,158 | 87,321,395 | 98.13 | 63,413,638 | 72.62% |
| 4 | ZT4_rep2 | 100,754,314 | 100,727,654 | 98.21 | 73,690,375 | 73.16% |
| 5 | ZT8_rep1 | 97,511,686 | 97,485,539 | 98.24 | 69,267,392 | 71.05% |
| 6 | ZT8_rep2 | 96,598,065 | 96,572,858 | 98.23 | 70,045,246 | 72.53% |
| 7 | ZT12_rep1 | 95,002,635 | 94,977,251 | 98.25 | 69,348,349 | 73.02% |
| 8 | ZT12_rep2 | 96,254,940 | 96,229,049 | 98.28 | 70,228,180 | 72.98% |
| 9 | ZT16_rep1 | 92,587,264 | 92,562,453 | 98.27 | 66,796,081 | 72.16% |
| 10 | ZT16_rep2 | 96,368,659 | 96,342,993 | 98.25 | 68,985,090 | 71.6% |
| 11 | ZT20_rep1 | 98,509,742 | 98,483,494 | 98.25 | 70,150,697 | 71.23% |
| 12 | ZT20_rep2 | 94,190,011 | 94,164,614 | 98.29 | 67,805,050 | 72.01% |
| 13 | ZT24_rep1 | 100,934,518 | 100,909,158 | 98.26 | 72,773,028 | 72.12% |
| 14 | ZT24_rep2 | 108,544,184 | 108,517,103 | 98.28 | 77,988,632 | 71.87% |
| AGI | Alias | Description | ID | PER | LAG | AMP |
|---|---|---|---|---|---|---|
| AT1G09570.1 | FHY2,FRE1,HY8,PHYA | phytochrome A | Bra020013 | 24.00 | 2.00 | 3.57 |
| Bra031672 | 18.00 | NA | 0.06 | |||
| AT2G18790.1 | HY3,OOP1,PHYB | phytochrome B | Bra001650 | 24.00 | 13.00 | 0.07 |
| Bra022192 | 22.00 | 7.15 | 3.58 | |||
| AT4G08920.1 | ATCRY1,BLU1,CRY1,HY4,OOP2 | cryptochrome 1 | Bra037880 | 24.00 | 2.00 | 58.64 |
| AT1G04400.1 | AT-PHH1,ATCRY2,CRY2,FHA,PHH1 | cryptochrome 2 | Bra015313 | 12.00 | 0.00 | 9.70 |
| Bra030568 | 10.00 | NA | 0.00 | |||
| AT2G32950.1 | ATCOP1,COP1,DET340,EMB168,FUS1 | transducin/WD40 repeat-like superfamily protein | Bra005541 | 20.00 | 7.00 | 5.27 |
| Bra021818 | 18.00 | NA | 0.04 | |||
| AT1G09530.1 | PAP3,PIF3,POC1 | phytochrome interacting factor 3 | Bra020017 | 24.00 | 5.00 | 3.75 |
| Bra031668 | 24.00 | 5.00 | 2.83 | |||
| AT1G22770.1 | FB,GI | gigantea protein (GI) | Bra024536 | 24.00 | 8.00 | 8.20 |
| AT2G25930.1 | ELF3,PYK20 | hydroxyproline-rich glycoprotein family protein | Bra007774 | 24.00 | 18.00 | 4.14 |
| Bra034284 | 20.00 | 0.00 | 2.22 | |||
| AT2G40080.1 | ELF4 | Protein of unknown function (DUF1313) | Bra000165 | 24.00 | 13.00 | 13.00 |
| Bra004991 | 24.00 | 14.00 | 19.58 | |||
| Bra017035 | 20.80 | 12.58 | 20.58 | |||
| AT5G02840.1 | LCL1(RVE4) | LHY/CCA1-like 1 | Bra005751 | 24.00 | 0.00 | 14.20 |
| Bra005754 | 22.00 | 19.07 | 0.10 | |||
| Bra009562 | 22.00 | 0.00 | 10.42 | |||
| AT3G09600.1 | RVE8 | homeodomain-like superfamily protein | Bra029778 | 24.00 | 2.00 | 19.94 |
| Bra034074 | 24.00 | 2.00 | 10.72 | |||
| AT3G46640.1 | LUX,PCL1 | homeodomain-like superfamily protein | Bra018204 | 24.00 | 16.00 | 4.89 |
| Bra033809 | 22.00 | 14.12 | 8.36 | |||
| AT2G18915.2 | ADO2,LKP2 | LOV KELCH protein 2 | Bra038830 | 24.00 | 3.00 | 1.58 |
| Bra038831 | 24.00 | 2.00 | 1.29 | |||
| Bra038832 | 22.67 | 11.96 | 5.81 | |||
| AT1G01060.1 AT1G01060.4 | LHY,LHY1 | homeodomain-like superfamily protein | Bra030496 | 24.00 | 1.00 | 34.47 |
| Bra033291 | 24.00 | 0.00 | 37.84 | |||
| AT2G46830.1 | CCA1 | circadian clock associated 1 | Bra004503 | 24.00 | 2.00 | 33.20 |
| AT5G61380.1 | APRR1,AtTOC1,PRR1,TOC1 | CCT motif -containing response regulator protein | Bra012964 | 22.67 | 11.96 | 15.09 |
| Bra035933 | 24.00 | 12.00 | 6.58 | |||
| AT5G60100.1 AT5G60100.2 | APRR3,PRR3 | pseudo-response regulator 3 | Bra002512 | 20.00 | 10.00 | 22.85 |
| Bra020263 | 18.00 | NA | NA | |||
| AT5G24470.1 | APRR5,PRR5 | pseudo-response regulator 5 | Bra009768 | 20.00 | 10.00 | 1.19 |
| Bra029407 | 20.00 | 10.00 | 3.28 | |||
| Bra036517 | 20.00 | 8.00 | 2.12 | |||
| AT5G02810.1 | APRR7,PRR7 | pseudo-response regulator 7 | Bra009565 | 24.00 | 8.00 | 25.25 |
| Bra028861 | 22.00 | 10.08 | 40.83 | |||
| AT2G46790.1 | APRR9,PRR9,TL1 | pseudo-response regulator 9 | Bra004507 | 24.00 | 6.00 | 0.11 |
| Bra040484 | 22.67 | 0.62 | 12.55 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.A.; Shim, D.; Kumari, S.; Jung, H.-e.; Jung, K.-H.; Jeong, H.; Kim, W.-Y.; Lee, S.I.; Jeong, M.-J. Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage. Genes 2019, 10, 130. https://doi.org/10.3390/genes10020130
Kim JA, Shim D, Kumari S, Jung H-e, Jung K-H, Jeong H, Kim W-Y, Lee SI, Jeong M-J. Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage. Genes. 2019; 10(2):130. https://doi.org/10.3390/genes10020130
Chicago/Turabian StyleKim, Jin A., Donghwan Shim, Shipra Kumari, Ha-eun Jung, Ki-Hong Jung, Heesu Jeong, Woe-Yeon Kim, Soo In Lee, and Mi-Jeong Jeong. 2019. "Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage" Genes 10, no. 2: 130. https://doi.org/10.3390/genes10020130
APA StyleKim, J. A., Shim, D., Kumari, S., Jung, H.-e., Jung, K.-H., Jeong, H., Kim, W.-Y., Lee, S. I., & Jeong, M.-J. (2019). Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage. Genes, 10(2), 130. https://doi.org/10.3390/genes10020130





