On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway
Abstract
:1. Introduction
2. Licensing and Activation of Replication Origins
3. Intra-S Phase Checkpoint Activation and Response
3.1. Replicative Stresses
3.2. Intra-S Phase Checkpoint Activation and Fork Stability
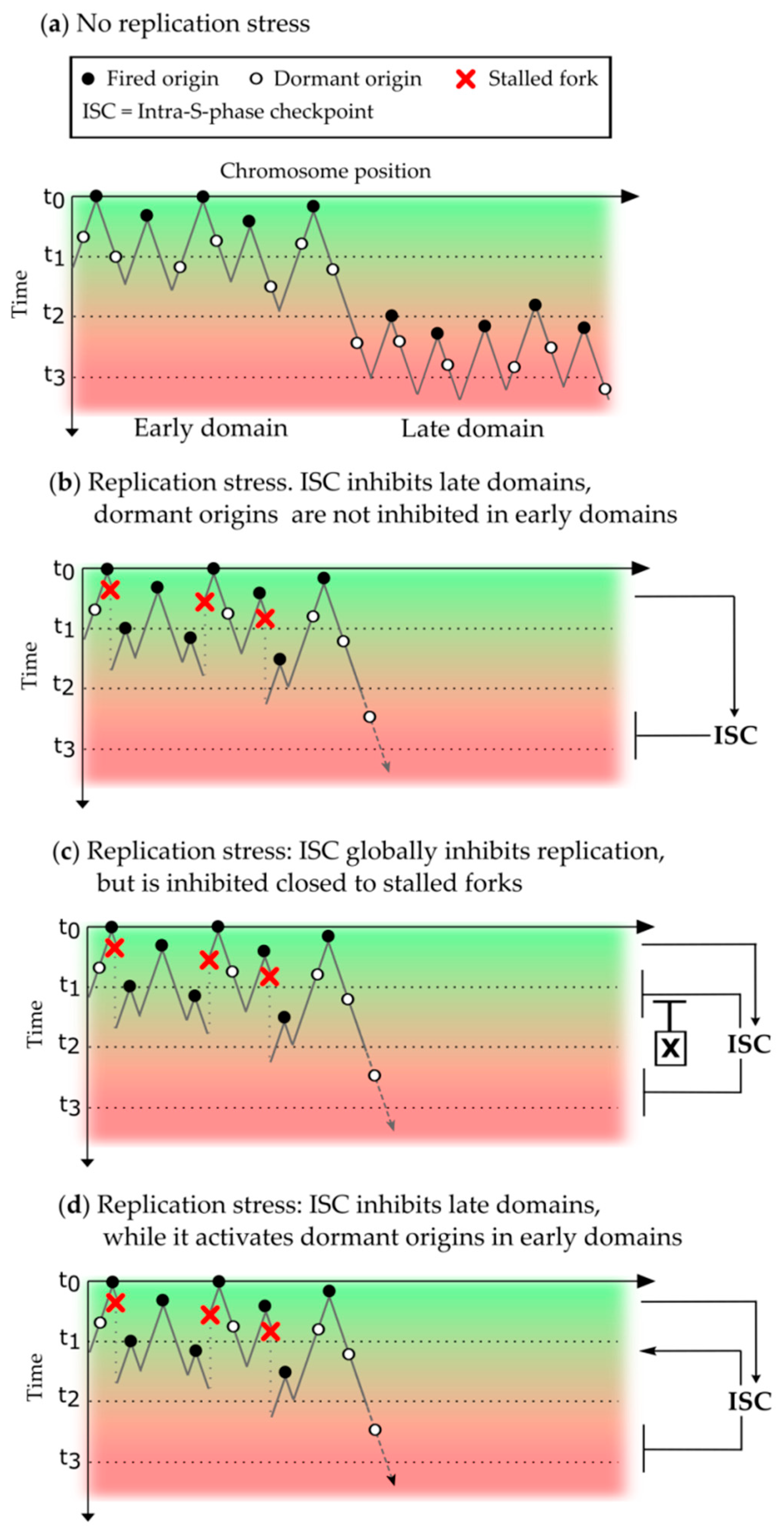
3.3. Inhibition of the Replication Program by the Intra-S Phase Checkpoint
3.4. Molecular Mechanisms of Checkpoint Dependent Origin Inhibition
3.4.1. In Response to Exogenous Stress
3.4.2. In Response to Endogenous Stress
4. Intra-S Phase Checkpoint Recovery and Adaptation
5. DNA Replication and Intra-S Phase Checkpoint Response in Rapid Cell Cycles During Early Development
6. Numerical Simulations and Models of the Spatio-Temporal Program of DNA Replication
6.1. Different Approaches to Analyze the Spatio-Temporal Replication Program
6.2. Different Approaches to Analyze the Intra-S Phase Checkpoint Action on the Replication Program
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 9-1-1 | Rad9-Rad1-Hus1 |
| APC | Anaphase-promoting complex |
| APH | Aphidicolin |
| AT | Ataxia telangiectasia |
| ATM | Ataxia telangiectasia mutated |
| ATR | Ataxia telangiectasia and Rad3-related protein |
| BrdU | Bromodeoxyuridine |
| Cdc | Cell division cycle |
| CDK | Cyclin-dependent kinase |
| Cds1 | Checking DNA synthesis 1 |
| Cdt1 | Chromatin licensing and DNA replication factor 1 |
| Chk1 | Checkpoint kinase 1 |
| Chk2 | Checkpoint kinase 2 |
| CMG | CDC45-MCM-GINS |
| Dbf4 | Dumbbell factor 4 |
| Dpb11 | DNA polymerase B-binding protein subunit 11 |
| DDK | Dbf4-dependent kinase |
| DNA | Deoxyribonucleic acid |
| dNTP | Deoxyribonucleotides |
| Drf1 | Dumbell-related factor 1 |
| dsDNA | Double stranded DNA |
| FANCI | Fanconi anemia complementation group I |
| GINS | Go-ichi-ni-san |
| HU | Hydroxyurea |
| kb | Kilobase |
| KJMA | Kolmogorov-Johnson-Mehl-Avrami |
| MBT | Mid-blastula transition |
| MCM | Minichromosome maintenance |
| Mec1 | Mitosis entry checkpoint 1 |
| MLL | Mixed-lineage leukemia |
| MMS | Methyl methane sulphonate |
| Mrc1 | Mannose receptor C-type 1 |
| ORC | Origin recognition complex |
| Plk1 | Polo-like kinase 1 |
| PP | Protein phosphatase |
| Pre-IC | Pre-initiation complex |
| Pre-RC | Pre-recognition complex |
| Rad | Radiation sensitive |
| RFC | Replication factor C |
| RIF1 | Rap1-interacting factor 1 |
| RNA | Ribonucleic acid |
| RPA | Replication protein A |
| RT | Replication timing |
| SCF | Skp, Cullin, F-box containing |
| Sld | Synthetic lethal with Dpb11 |
| ssDNA | Single stranded DNA |
| Ticrr | TopBP1 interacting checkpoint and replication regulator |
| TLS | Translesion synthesis |
| TopBP1 | Topoisomerase II binding protein 1 |
References
- Machida, Y.J.; Hamlin, J.L.; Dutta, A. Right place, right time, and only once: replication initiation in metazoans. Cell 2005, 123, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O.; Marheineke, K.; Goldar, A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. BioEssays 2003, 25, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Audit, B.; Baker, A.; Chen, C.-L.; Rappailles, A.; Guilbaud, G.; Julienne, H.; Goldar, A.; d’Aubenton-Carafa, Y.; Hyrien, O.; Thermes, C.; et al. Multiscale analysis of genome-wide replication timing profiles using a wavelet-based signal-processing algorithm. Nat. Protoc. 2013, 8, 98–110. [Google Scholar] [CrossRef]
- Eshaghi, M.; Karuturi, R.K.M.; Li, J.; Chu, Z.; Liu, E.T.; Liu, J. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS ONE 2007, 2, e722. [Google Scholar] [CrossRef] [PubMed]
- Heichinger, C.; Penkett, C.J.; Bähler, J.; Nurse, P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006, 25, 5171–5179. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K.; Winzeler, E.A.; Collingwood, D.; Hunt, S.; Wodicka, L.; Conway, A.; Lockhart, D.J.; Davis, R.W.; Brewer, B.J.; Fangman, W.L. Replication dynamics of the yeast genome. Science 2001, 294, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K.; Brewer, B.J.; Fangman, W.L. Cell cycle-dependent establishment of a late replication program. Science 1997, 276, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Lukas, J.; Lukas, C.; Bartek, J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair 2004, 3, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 2001, 13, 738–747. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Weinert, T.A. Checkpoints: controls that ensure the order of cell cycle events. Science 1989, 246, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, R.; Cimprich, K. The ATR pathway: Fine-tuning the fork. DNA Repair 2007, 6, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Galanti, L.; Pfander, B. Right time, right place—DNA damage and DNA replication checkpoints collectively safeguard S phase. EMBO J. 2018, 37, e100681. [Google Scholar] [CrossRef] [PubMed]
- Goldar, A.; Marsolier-Kergoat, M.-C.; Hyrien, O. Universal Temporal Profile of Replication Origin Activation in Eukaryotes. PLoS ONE 2009, 4, e5899. [Google Scholar] [CrossRef] [PubMed]
- Goldar, A.; Labit, H.; Marheineke, K.; Hyrien, O. A Dynamic Stochastic Model for DNA Replication Initiation in Early Embryos. PLoS ONE 2008, 3, e2919. [Google Scholar] [CrossRef] [PubMed]
- Arbona, J.-M.; Goldar, A.; Hyrien, O.; Arneodo, A.; Audit, B. The eukaryotic bell-shaped temporal rate of DNA replication origin firing emanates from a balance between origin activation and passivation. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Goldar, A.; Arneodo, A.; Audit, B.; Argoul, F.; Rappailles, A.; Guilbaud, G.; Petryk, N.; Kahli, M.; Hyrien, O. Deciphering DNA replication dynamics in eukaryotic cell populations in relation with their averaged chromatin conformations. Sci. Rep. 2016, 6, 22469. [Google Scholar] [CrossRef] [PubMed]
- Gindin, Y.; Valenzuela, M.S.; Aladjem, M.I.; Meltzer, P.S.; Bilke, S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol. Syst. Biol. 2014, 10, 722. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.G.; Bechhoefer, J. Control of DNA Replication by Anomalous Reaction-Diffusion Kinetics. Phys. Rev. Lett. 2009, 102, 158104. [Google Scholar] [CrossRef] [PubMed]
- Segurado, M.; Diffley, J.F.X. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008, 22, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Brocas, C.; Charbonnier, J.-B.; Dhérin, C.; Gangloff, S.; Maloisel, L. Stable interactions between DNA polymerase δ catalytic and structural subunits are essential for efficient DNA repair. DNA Repair 2010, 9, 1098–1111. [Google Scholar] [CrossRef]
- Saldivar, J.C.; Cortez, D.; Cimprich, K.A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Iyer, D.; Rhind, N. The Intra-S Checkpoint Responses to DNA Damage. Genes 2017, 8, 74. [Google Scholar] [CrossRef]
- Shaltiel, I.A.; Krenning, L.; Bruinsma, W.; Medema, R.H. The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015, 128, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, M.; Branzei, D. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cell. Mol. Life Sci. 2017, 74, 2361–2380. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O. Peaks cloaked in the mist: the landscape of mammalian replication origins. J. Cell Biol. 2015, 208, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic Origin-Dependent DNA Replication In Vitro Reveals Sequential Action of DDK and S-CDK Kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Watase, G.; Takisawa, H.; Kanemaki, M.T. Mcm10 Plays a Role in Functioning of the Eukaryotic Replicative DNA Helicase, Cdc45-Mcm-GINS. Curr. Biol. 2012, 22, 343–349. [Google Scholar] [CrossRef]
- Simon, A.C.; Zhou, J.C.; Perera, R.L.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinković, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef]
- Herrick, J.; Stanislawski, P.; Hyrien, O.; Bensimon, A. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J. Mol. Biol. 2000, 300, 1133–1142. [Google Scholar] [CrossRef]
- Zegerman, P.; Diffley, J.F.X. DNA replication as a target of the DNA damage checkpoint. DNA Repair 2009, 8, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.L. (Ed.) The Initiation of DNA Replication in Eukaryotes; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24694-9. [Google Scholar]
- Rhind, N.; Gilbert, D.M. DNA Replication Timing. Cold Spring Harb. Perspect. Biol. 2013, 5, a010132. [Google Scholar] [CrossRef]
- Bechhoefer, J.; Rhind, N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012, 28, 374–381. [Google Scholar] [CrossRef]
- Mantiero, D.; Mackenzie, A.; Donaldson, A.; Zegerman, P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011, 30, 4805–4814. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Vindigni, A. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 2016, 23, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Hennion, M.; Arbona, J.-M.; Cruaud, C.; Proux, F.; Tallec, B.L.; Novikova, E.; Engelen, S.; Lemainque, A.; Audit, B.; Hyrien, O. Mapping DNA replication with nanopore sequencing. bioRxiv 2018. [Google Scholar] [CrossRef]
- Nieduszynski, C. Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. bioRxiv 2018. [Google Scholar] [CrossRef]
- Sorensen, C.S.; Syljuasen, R.G. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012, 40, 477–486. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Cabrera, E.; Freire, R.; Gillespie, D.A. Claspin—Checkpoint adaptor and DNA replication factor. FEBS J. 2018, 286, 441–455. [Google Scholar] [CrossRef]
- Ibarra, A.; Schwob, E.; Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 2008, 105, 8956–8961. [Google Scholar] [CrossRef]
- Ge, X.Q.; Jackson, D.A.; Blow, J.J. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007, 21, 3331–3341. [Google Scholar] [CrossRef]
- Woodward, A.M.; Göhler, T.; Luciani, M.G.; Oehlmann, M.; Ge, X.; Gartner, A.; Jackson, D.A.; Blow, J.J. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006, 173, 673–683. [Google Scholar] [CrossRef]
- Lucas, I.; Chevrier-Miller, M.; Sogo, J.M.; Hyrien, O. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 2000, 296, 769–786. [Google Scholar] [CrossRef] [PubMed]
- Anglana, M.; Apiou, F.; Bensimon, A.; Debatisse, M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 2003, 114, 385–394. [Google Scholar] [CrossRef]
- Ge, X.Q.; Blow, J.J. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 2010, 191, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Santocanale, C.; Sharma, K.; Diffley, J.F.X. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999, 13, 2360–2364. [Google Scholar] [CrossRef] [PubMed]
- Vujcic, M.; Miller, C.A.; Kowalski, D. Activation of Silent Replication Origins at Autonomously Replicating Sequence Elements near the HML Locus in Budding Yeast. Mol. Cell. Biol. 1999, 19, 6098–6109. [Google Scholar] [CrossRef]
- Siddiqui, K.; On, K.F.; Diffley, J.F.X. Regulating DNA Replication in Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef]
- Lucca, C.; Vanoli, F.; Cotta-Ramusino, C.; Pellicioli, A.; Liberi, G.; Haber, J.; Foiani, M. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 2004, 23, 1206–1213. [Google Scholar] [CrossRef]
- Cobb, J.A.; Bjergbaek, L.; Shimada, K.; Frei, C.; Gasser, S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003, 22, 4325–4336. [Google Scholar] [CrossRef]
- Painter, R.B.; Young, B.R. Radiosensitivity in ataxia-telangiectasia: A new explanation. Proc. Natl. Acad. Sci. USA 1980, 77, 7315–7317. [Google Scholar] [CrossRef] [PubMed]
- Paulovich, A.G.; Hartwell, L.H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in Response to DNA damage. Cell 1995, 82, 841–847. [Google Scholar] [CrossRef]
- Santocanale, C.; Diffley, J.F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 1998, 395, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Shirahige, K.; Hori, Y.; Shiraishi, K.; Yamashita, M.; Takahashi, K.; Obuse, C.; Tsurimoto, T.; Yoshikawa, H. Regulation of DNA-replication origins during cell-cycle progression. Nature 1998, 395, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Collingwood, D.; Boeck, M.E.; Fox, L.A.; Alvino, G.M.; Fangman, W.L.; Raghuraman, M.K.; Brewer, B.J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006, 8, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Alvino, G.M.; Collingwood, D.; Murphy, J.M.; Delrow, J.; Brewer, B.J.; Raghuraman, M.K. Replication in Hydroxyurea: It’s a Matter of Time. Mol. Cell. Biol. 2007, 27, 6396–6406. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K.; Brewer, B.J. Molecular analysis of the replication program in unicellular model organisms. Chromosome Res. 2010, 18, 19–34. [Google Scholar] [CrossRef]
- Crabbé, L.; Thomas, A.; Pantesco, V.; De Vos, J.; Pasero, P.; Lengronne, A. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat. Struct. Mol. Biol. 2010, 17, 1391–1397. [Google Scholar] [CrossRef]
- Larner, J.M.; Lee, H.; Dijkwel, P.A.; Little, R.D.; Schildkraut, C.L.; Hamlin, J.L. Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucleic Acids Res. 1999, 27, 803–809. [Google Scholar] [CrossRef]
- Dimitrova, D.S.; Gilbert, D.M. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2000, 2, 686–694. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Petermann, E.; Gillespie, D.A.F.; Caldecott, K.W.; Jackson, D.A. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007, 26, 2719–2731. [Google Scholar] [CrossRef] [PubMed]
- Karnani, N.; Dutta, A. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 2011, 25, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Marheineke, K.; Hyrien, O. Control of replication origin density and firing time in Xenopus egg extracts: Role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 2004, 279, 28071–28081. [Google Scholar] [CrossRef] [PubMed]
- Shechter, D.; Costanzo, V.; Gautier, J. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 2004, 6, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Luciani, M.G. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 2004, 117, 6019–6030. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.M.; Michael, W.M. Control of DNA Replication by the Nucleus/Cytoplasm Ratio in Xenopus. J. Biol. Chem. 2013, 288, 29382–29393. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Kumagai, A.; Schlacher, K.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Interaction of Chk1 with Treslin Negatively Regulates the Initiation of Chromosomal DNA Replication. Mol. Cell 2015, 57, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Platel, M.; Goldar, A.; Wiggins, J.M.; Barbosa, P.; Libeau, P.; Priam, P.; Narassimprakash, H.; Grodzenski, X.; Marheineke, K. Tight Chk1 levels control replication cluster activation in Xenopus. PLoS ONE 2015, 10, e0129090. [Google Scholar] [CrossRef] [PubMed]
- Yekezare, M.; Gómez-González, B.; Diffley, J.F.X. Controlling DNA replication origins in response to DNA damage—Inhibit globally, activate locally. J. Cell Sci. 2013, 126, 1297–1306. [Google Scholar] [CrossRef]
- Trenz, K.; Errico, A.; Costanzo, V. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008, 27, 876–885. [Google Scholar] [CrossRef]
- Jares, P.; Blow, J.J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000, 14, 1528–1540. [Google Scholar] [PubMed]
- Walter, J.C. Evidence for Sequential Action of cdc7 and cdk2 Protein Kinases during Initiation of DNA Replication in Xenopus Egg Extracts. J. Biol. Chem. 2000, 275, 39773–39778. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Kawasaki, Y.; Young, M.R.; Kihara, M.; Sugino, A.; Tye, B.K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997, 11, 3365–3374. [Google Scholar] [CrossRef] [PubMed]
- Sheu, Y.-J.; Stillman, B. Cdc7-Dbf4 Phosphorylates MCM Proteins via a Docking Site-Mediated Mechanism to Promote S Phase Progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Abid Ali, F.; Douglas, M.E.; Locke, J.; Pye, V.E.; Nans, A.; Diffley, J.F.X.; Costa, A. Cryo-EM structure of a licensed DNA replication origin. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Treslin Collaborates with TopBP1 in Triggering the Initiation of DNA Replication. Cell 2010, 140, 349–359. [Google Scholar] [CrossRef]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 2011, 193, 995–1007. [Google Scholar] [CrossRef]
- Boos, D.; Sanchez-Pulido, L.; Rappas, M.; Pearl, L.H.; Oliver, A.W.; Ponting, C.P.; Diffley, J.F.X. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Curr. Biol. 2011, 21, 1152–1157. [Google Scholar] [CrossRef]
- Sangrithi, M.N.; Bernal, J.A.; Madine, M.; Philpott, A.; Lee, J.; Dunphy, W.G.; Venkitaraman, A.R. Initiation of DNA Replication Requires the RECQL4 Protein Mutated in Rothmund-Thomson Syndrome. Cell 2005, 121, 887–898. [Google Scholar] [CrossRef]
- Matsuno, K.; Kumano, M.; Kubota, Y.; Hashimoto, Y.; Takisawa, H. The N-Terminal Noncatalytic Region of Xenopus RecQ4 Is Required for Chromatin Binding of DNA Polymerase in the Initiation of DNA Replication. Mol. Cell. Biol. 2006, 26, 4843–4852. [Google Scholar] [CrossRef] [PubMed]
- Karlsson-Rosenthal, C.; Millar, J.B.A. Cdc25: Mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006, 16, 285–292. [Google Scholar] [CrossRef]
- Donzelli, M.; Draetta, G.F. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003, 4, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Goto, H.; Enomoto, M.; Tomono, Y.; Kiyono, T.; Inagaki, M. 14-3-3γ mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010, 29, 2802–2812. [Google Scholar] [CrossRef] [PubMed]
- Melixetian, M.; Klein, D.K.; Sørensen, C.S.; Helin, K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 2009, 11, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Hassepass, I.; Voit, R.; Hoffmann, I. Phosphorylation at Serine 75 Is Required for UV-mediated Degradation of Human Cdc25A Phosphatase at the S-phase Checkpoint. J. Biol. Chem. 2003, 278, 29824–29829. [Google Scholar] [CrossRef]
- Busino, L.; Chiesa, M.; Draetta, G.F.; Donzelli, M. Cdc25A phosphatase: Combinatorial phosphorylation, ubiquitylation and proteolysis. Oncogene 2004, 23, 2050–2056. [Google Scholar] [CrossRef]
- Busino, L.; Donzelli, M.; Chiesa, M.; Guardavaccaro, D.; Ganoth, D.; Dorrello, N.V.; Hershko, A.; Pagano, M.; Draetta, G.F. Degradation of Cdc25A by b-TrCP during S phase and in response to DNA damage. Nature 2003, 426, 87–91. [Google Scholar] [CrossRef]
- Jin, J.; Shirogane, T.; Xu, L.; Nalepa, G.; Qin, J.; Elledge, S.J.; Harper, J.W. SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003, 17, 3062–3074. [Google Scholar] [CrossRef]
- Mailand, N. Rapid Destruction of Human Cdc25A in Response to DNA Damage. Science 2000, 288, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Goloudina, A.; Yamaguchi, H.; Chervyakova, D.B.; Appella, E.; Fornace, A.J., Jr.; Bulavin, D.V. Regulation of Human Cdc25A Stability by Serine 75 Phosphorylation Is Not Sufficient to Activate a S-phase Checkpoint. Cell Cycle 2003, 2, 471–476. [Google Scholar] [CrossRef]
- Uto, K.; Inoue, D.; Shimuta, K.; Nakajo, N.; Sagata, N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004, 23, 3386–3396. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-S.; Ryan, C.E.; Piwnica-Worms, H. Chk1 Kinase Negatively Regulates Mitotic Function of Cdc25A Phosphatase through 14-3-3 Binding. Mol. Cell. Biol. 2003, 23, 7488–7497. [Google Scholar] [CrossRef]
- Lopez-Mosqueda, J.; Maas, N.L.; Jonsson, Z.O.; DeFazio-Eli, L.G.; Wohlschlegel, J.; Toczyski, D.P. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 2010, 467, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 2010, 467, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Can, G.; Kauerhof, A.C.; Macak, D.; Zegerman, P. Helicase Subunit Cdc45 Targets the Checkpoint Kinase Rad53 to Both Replication Initiation and Elongation Complexes after Fork Stalling. Mol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Pasero, P.; Duncker, B.P.; Schwob, E.; Gasser, S.M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 1999, 13, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.; Stillman, B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999, 18, 5334–5346. [Google Scholar] [CrossRef] [PubMed]
- Kihara, M.; Nakai, W.; Asano, S.; Suzuki, A.; Kitada, K.; Kawasaki, Y.; Johnston, L.H.; Sugino, A. Characterization of the Yeast Cdc7p/Dbf4p Complex Purified from Insect Cells: ITS PROTEIN KINASE ACTIVITY IS REGULATED BY Rad53p. J. Biol. Chem. 2000, 275, 35051–35062. [Google Scholar] [CrossRef] [PubMed]
- Sheu, Y.-J.; Kinney, J.B.; Stillman, B. Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res. 2016, 26, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, V.; Shechter, D.; Lupardus, P.J.; Cimprich, K.A.; Gottesman, M.; Gautier, J. An ATR- and Cdc7-Dependent DNA Damage Checkpoint that Inhibits Initiation of DNA Replication. Mol. Cell 2003, 11, 203–213. [Google Scholar] [CrossRef]
- Heffernan, T.P.; Ünsal-Kaçmaz, K.; Heinloth, A.N.; Simpson, D.A.; Paules, R.S.; Sancar, A.; Cordeiro-Stone, M.; Kaufmann, W.K. Cdc7/Dbf4 and the Human S Checkpoint Response to UVC. J. Biol. Chem. 2007, 282, 9458–9468. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Poh, W.T.; Chadha, G.S.; Gillespie, P.J.; Kaldis, P.; Blow, J.J. Xenopus Cdc7 executes its essential function early in S phase and is counteracted by checkpoint-regulated protein phosphatase 1. Open Biol. 2014, 4. [Google Scholar] [CrossRef]
- Tsuji, T.; Lau, E.; Chiang, G.G.; Jiang, W. The role of Dbf4/Drf1-dependent kinase Cdc7 (Ddk) in DNA damage checkpoint control. Mol. Cell 2008, 32, 862–869. [Google Scholar] [CrossRef]
- Yanow, S.K.; Gold, D.A.; Yoo, H.Y.; Dunphy, W.G. Xenopus Drf1, a Regulator of Cdc7, Displays Checkpoint-dependent Accumulation on Chromatin during an S-phase Arrest. J. Biol. Chem. 2003, 278, 41083–41092. [Google Scholar] [CrossRef]
- Tenca, P.; Brotherton, D.; Montagnoli, A.; Rainoldi, S.; Albanese, C.; Santocanale, C. Cdc7 Is an Active Kinase in Human Cancer Cells Undergoing Replication Stress. J. Biol. Chem. 2007, 282, 208–215. [Google Scholar] [CrossRef]
- Yamada, M.; Watanabe, K.; Mistrik, M.; Vesela, E.; Protivankova, I.; Mailand, N.; Lee, M.; Masai, H.; Lukas, J.; Bartek, J. ATR–Chk1–APC/CCdh1-dependent stabilization of Cdc7–ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 2013, 27, 2459–2472. [Google Scholar] [CrossRef]
- Lee, A.Y.-L.; Chiba, T.; Truong, L.N.; Cheng, A.N.; Do, J.; Cho, M.J.; Chen, L.; Wu, X. Dbf4 Is Direct Downstream Target of Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad3-related (ATR) Protein to Regulate Intra-S-phase Checkpoint. J. Biol. Chem. 2012, 287, 2531–2543. [Google Scholar] [CrossRef]
- Yamada, M.; Masai, H.; Bartek, J. Regulation and roles of Cdc7 kinase under replication stress. Cell Cycle 2014, 13, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Takeda, S.; Kumar, R.; Westergard, T.D.; Brown, E.J.; Pandita, T.K.; Cheng, E.H.-Y.; Hsieh, J.J.-D. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature 2010, 467, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Räschle, M.; Knipscheer, P.; Enoiu, M.; Angelov, T.; Sun, J.; Griffith, J.D.; Ellenberger, T.E.; Schärer, O.D.; Walter, J.C. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell 2008, 134, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Zimmer, J.; Tarsounas, M. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016, 35, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Lossaint, G.; Larroque, M.; Ribeyre, C.; Bec, N.; Larroque, C.; Décaillet, C.; Gari, K.; Constantinou, A. FANCD2 Binds MCM Proteins and Controls Replisome Function upon Activation of S Phase Checkpoint Signaling. Mol. Cell 2013, 51, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Jones, M.J.K.; Yin, Y.; Crist, S.B.; Colnaghi, L.; Sims, R.J.; Rothenberg, E.; Jallepalli, P.V.; Huang, T.T. ATR-Mediated Phosphorylation of FANCI Regulates Dormant Origin Firing in Response to Replication Stress. Mol. Cell 2015, 58, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.L.; Yeo, J.E.; Lee, E.-A.; Kan, Y.; Raghunandan, M.; Wiek, C.; Hanenberg, H.; Schärer, O.D.; Hendrickson, E.A.; Sobeck, A. FANCI and FANCD2 have common as well as independent functions during the cellular replication stress response. Nucleic Acids Res. 2017, 45, 11837–11857. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Kosiyatrakul, S.T.; Boisvert, R.A.; Herrera-Moyano, E.; García-Rubio, M.L.; Gerhardt, J.; Vuono, E.A.; Owen, N.; Yan, Z.; Olson, S.; et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol. Cell 2016, 64, 388–404. [Google Scholar] [CrossRef]
- Syljuasen, R.G.; Sorensen, C.S.; Hansen, L.T.; Fugger, K.; Lundin, C.; Johansson, F.; Helleday, T.; Sehested, M.; Lukas, J.; Bartek, J. Inhibition of Human Chk1 Causes Increased Initiation of DNA Replication, Phosphorylation of ATR Targets, and DNA Breakage. Mol. Cell. Biol. 2005, 25, 3553–3562. [Google Scholar] [CrossRef]
- Miao, H. Regulation of Cellular and SV40 Virus Origins of Replication by Chk1-dependent Intrinsic and UVC Radiation-induced Checkpoints. J. Biol. Chem. 2003, 278, 4295–4304. [Google Scholar] [CrossRef]
- Scorah, J.; McGowan, C.H. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle 2009, 8, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Maya-Mendoza, A.; Zachos, G.; Gillespie, D.A.F.; Jackson, D.A.; Caldecott, K.W. Chk1 Requirement for High Global Rates of Replication Fork Progression during Normal Vertebrate S Phase. Mol. Cell. Biol. 2006, 26, 3319–3326. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Woodcock, M.; Helleday, T. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. USA 2010, 107, 16090–16095. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, C.S.; Syljuåsen, R.G.; Lukas, J.; Bartek, J. ATR, Claspin and the Rad9-Rad1-Hus1 Complex Regulate Chk1 and Cdc25A in the Absence of DNA Damage. Cell Cycle 2004, 3, 939–943. [Google Scholar] [CrossRef]
- Sørensen, C.S.; Syljuåsen, R.G.; Falck, J.; Schroeder, T.; Rönnstrand, L.; Khanna, K.K.; Zhou, B.-B.; Bartek, J.; Lukas, J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 2003, 3, 247–258. [Google Scholar] [CrossRef]
- Zhao, H.; Watkins, J.L.; Piwnica-Worms, H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. USA 2002, 99, 14795–14800. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Mayca Pozo, F.; Wisotsky, J.N.; Wang, B.; Jacobberger, J.W.; Zhang, Y. Phosphorylation of Minichromosome Maintenance 3 (MCM3) by Checkpoint Kinase 1 (Chk1) Negatively Regulates DNA Replication and Checkpoint Activation. J. Biol. Chem. 2015, 290, 12370–12378. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, T.; Hood, B.; Schamus, S.; O’Connor, M.J.; Conrads, T.P.; Bakkenist, C.J. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Guillemain, G.; Ma, E.; Mauger, S.; Miron, S.; Thai, R.; Guerois, R.; Ochsenbein, F.; Marsolier-Kergoat, M.-C. Mechanisms of Checkpoint Kinase Rad53 Inactivation after a Double-Strand Break in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3378–3389. [Google Scholar] [CrossRef]
- Leroy, C.; Lee, S.E.; Vaze, M.B.; Ochsenbien, F.; Guerois, R.; Haber, J.E.; Marsolier-Kergoat, M.-C. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 2003, 11, 827–835. [Google Scholar] [CrossRef]
- Leung-Pineda, V.; Ryan, C.E.; Piwnica-Worms, H. Phosphorylation of Chk1 by ATR Is Antagonized by a Chk1-Regulated Protein Phosphatase 2A Circuit. Mol. Cell. Biol. 2006, 26, 7529–7538. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Trastoy, M.; Berthonaud, V.; Chevalier, A.; Ducrot, C.; Marsolier-Kergoat, M.-C.; Mann, C.; Leteurtre, F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene 2007, 26, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B.M.; Szyjka, S.J.; Lis, E.T.; Bailey, A.O.; Yates, J.R.; Aparicio, O.M.; Romesberg, F.E. Pph3 Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA 2007, 104, 9290–9295. [Google Scholar]
- Travesa, A.; Duch, A.; Quintana, D.G. Distinct Phosphatases Mediate the Deactivation of the DNA Damage Checkpoint Kinase Rad53. J. Biol. Chem. 2008, 283, 17123–17130. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 2004, 117, 575–588. [Google Scholar] [CrossRef]
- Bennett, L.N.; Clarke, P.R. Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006, 580, 4176–4181. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Bartek, J.; Lukas, J. Destruction of Claspin by SCFβTrCP Restrains Chk1 Activation and Facilitates Recovery from Genotoxic Stress. Mol. Cell 2006, 23, 307–318. [Google Scholar] [CrossRef]
- Mamely, I.; van Vugt, M.A.; Smits, V.A.; Semple, J.I.; Lemmens, B.; Perrakis, A.; Medema, R.H.; Freire, R. Polo-like Kinase-1 Controls Proteasome-Dependent Degradation of Claspin during Checkpoint Recovery. Curr. Biol. 2006, 16, 1950–1955. [Google Scholar] [CrossRef]
- Peschiaroli, A.; Dorrello, N.V.; Guardavaccaro, D.; Venere, M.; Halazonetis, T.; Sherman, N.E.; Pagano, M. SCFβTrCP-Mediated Degradation of Claspin Regulates Recovery from the DNA Replication Checkpoint Response. Mol. Cell 2006, 23, 319–329. [Google Scholar] [CrossRef]
- Chaudhury, I.; Koepp, D. Recovery from the DNA Replication Checkpoint. Genes 2016, 7, 94. [Google Scholar] [CrossRef]
- Newport, J.; Dasso, M. On the coupling between DNA replication and mitosis. J. Cell Sci. Suppl. 1989, 12, 149–160. [Google Scholar] [CrossRef]
- Anderson, J.A.; Lewellyn, A.L.; Maller, J.L. Ionizing Radiation Induces Apoptosis and Elevates Cyclin Al-Cdk2 Activity Before but not after the Midblastula Transition in Xenopus. Mol. Biol. Cell 1997, 8, 12. [Google Scholar] [CrossRef]
- Clute, P.; Masui, Y. Microtubule Dependence of Chromosome Cycles inXenopus laevisBlastomeres under the Influence of a DNA Synthesis Inhibitor, Aphidicolin. Dev. Biol. 1997, 185, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Raff, J.W. Nuclear and cytoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited with aphidicolin. J. Cell Biol. 1988, 107, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, R.; Hunter, P.; Yager, T.D. Developmental Activation of the Capability to Undergo Checkpoint-Induced Apoptosis in the Early Zebrafish Embryo. Dev. Biol. 1999, 209, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Holway, A.H.; Kim, S.-H.; La Volpe, A.; Michael, W.M. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 2006, 172, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, P.H.; Stumpff, J.; Tin Su, T. Embryonic Cleavage Cycles: How Is a Mouse Like a Fly? Curr. Biol. 2004, 14, R35–R45. [Google Scholar] [CrossRef] [PubMed]
- Newport, J.; Kirschner, M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 1982, 30, 675–686. [Google Scholar] [CrossRef]
- Satoh, N. Timing mechanisms in early embryonic development. Differ. Res. Biol. Divers. 1982, 22, 156–163. [Google Scholar] [CrossRef]
- Kane, D.A.; Kimmel, C.B. The zebrafish midblastula transition. Development 1993, 119, 447–456. [Google Scholar]
- Masui, Y.; Wang, P. Cell cycle transition in early embryonic development of Xenopus laevis. Biol. Cell 1998, 90, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O.; Maric, C.; Méchali, M. Transition in specification of embryonic metazoan DNA replication origins. Science 1995, 270, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Newport, J.W. Regulation of replicon size in Xenopus egg extracts. Science 1997, 275, 993–995. [Google Scholar] [CrossRef] [PubMed]
- Kermi, C.; Lo Furno, E.; Maiorano, D. Regulation of DNA Replication in Early Embryonic Cleavages. Genes 2017, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.W.; Lewellyn, A.L.; Maller, J.L. The DNA damage checkpoint in embryonic cell cycles is dependent on the DNA-to-cytoplasmic ratio. Dev. Cell 2004, 7, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Lewellyn, A.L.; Maller, J.L. DNA damage signaling in early Xenopus embryos. Cell Cycle 2008, 7, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Shimuta, K.; Nakajo, N.; Uto, K.; Hayano, Y.; Okazaki, K.; Sagata, N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002, 21, 3694–3703. [Google Scholar] [CrossRef] [PubMed]
- Sibon, O.C.M.; Stevenson, V.A.; Theurkauf, W.E. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 1997, 388, 93–97. [Google Scholar] [CrossRef]
- Collart, C.; Smith, J.C.; Zegerman, P. Chk1 Inhibition of the Replication Factor Drf1 Guarantees Cell-Cycle Elongation at the Xenopus laevis Mid-blastula Transition. Dev. Cell 2017, 42, 82–96. [Google Scholar] [CrossRef]
- Collart, C.; Allen, G.E.; Bradshaw, C.R.; Smith, J.C.; Zegerman, P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 2013, 341, 893–896. [Google Scholar] [CrossRef]
- Collart, C.; Christov, C.P.; Smith, J.C.; Krude, T. The Midblastula Transition Defines the Onset of Y RNA-Dependent DNA Replication in Xenopus laevis. Mol. Cell. Biol. 2011, 31, 3857–3870. [Google Scholar] [CrossRef] [PubMed]
- Christov, C.P.; Dingwell, K.S.; Skehel, M.; Wilkes, H.S.; Sale, J.E.; Smith, J.C.; Krude, T. A NuRD Complex from Xenopus laevis Eggs Is Essential for DNA Replication during Early Embryogenesis. Cell Rep. 2018, 22, 2265–2278. [Google Scholar] [CrossRef] [PubMed]
- Amodeo, A.A.; Jukam, D.; Straight, A.F.; Skotheim, J.M. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. USA 2015, 112, E1086–E1095. [Google Scholar] [CrossRef] [PubMed]
- Seller, C.A.; O’Farrell, P.H. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol. 2018, 16, e2005687. [Google Scholar] [CrossRef] [PubMed]
- Karimi, K.; Fortriede, J.D.; Lotay, V.S.; Burns, K.A.; Wang, D.Z.; Fisher, M.E.; Pells, T.J.; James-Zorn, C.; Wang, Y.; Ponferrada, V.G.; et al. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018, 46, D861–D868. [Google Scholar] [CrossRef] [PubMed]
- Petersen, P.; Chou, D.M.; You, Z.; Hunter, T.; Walter, J.C.; Walter, G. Protein Phosphatase 2A Antagonizes ATM and ATR in a Cdk2- and Cdc7-Independent DNA Damage Checkpoint. Mol. Cell. Biol. 2006, 26, 1997–2011. [Google Scholar] [CrossRef] [PubMed]
- Kermi, C.; Prieto, S.; van der Laan, S.; Tsanov, N.; Recolin, B.; Uro-Coste, E.; Delisle, M.-B.; Maiorano, D. RAD18 Is a Maternal Limiting Factor Silencing the UV-Dependent DNA Damage Checkpoint in Xenopus Embryos. Dev. Cell 2015, 34, 364–372. [Google Scholar] [CrossRef]
- Blythe, S.A.; Wieschaus, E.F. Zygotic Genome Activation Triggers the DNA Replication Checkpoint at the Midblastula Transition. Cell 2015, 160, 1169–1181. [Google Scholar] [CrossRef]
- Herrick, J.; Jun, S.; Bechhoefer, J.; Bensimon, A. Kinetic model of DNA replication in eukaryotic organisms. J. Mol. Biol. 2002, 320, 741–750. [Google Scholar] [CrossRef]
- Jun, S.; Zhang, H.; Bechhoefer, J. Nucleation and growth in one dimension. I. The generalized Kolmogorov-Johnson-Mehl-Avrami model. Phys. Rev. E 2005, 71. [Google Scholar] [CrossRef]
- Jun, S.; Bechhoefer, J. Nucleation and growth in one dimension. II. Application to DNA replication kinetics. Phys. Rev. E 2005, 71. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.C.-H.; Bechhoefer, J. How Xenopus laevis embryos replicate reliably: Investigating the random-completion problem. Phys. Rev. E 2008, 78, 041917. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.C.-H.; Rhind, N.; Bechhoefer, J. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol. Syst. Biol. 2010, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- de Moura, A.P.S.; Retkute, R.; Hawkins, M.; Nieduszynski, C.A. Mathematical modelling of whole chromosome replication. Nucleic Acids Res. 2010, 38, 5623–5633. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Li, J.; Eshaghi, M.; Liu, J.; Karuturi, R.K.M. Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data. BMC Bioinform. 2010, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Retkute, R.; Nieduszynski, C.A.; de Moura, A. Dynamics of DNA Replication in Yeast. Phys. Rev. Lett. 2011, 107. [Google Scholar] [CrossRef]
- Baker, A.; Audit, B.; Yang, S.C.-H.; Bechhoefer, J.; Arneodo, A. Inferring Where and When Replication Initiates from Genome-Wide Replication Timing Data. Phys. Rev. Lett. 2012, 108, 268101. [Google Scholar] [CrossRef]
- Pichugina, T.; Sugawara, T.; Kaykov, A.; Schierding, W.; Masuda, K.; Uewaki, J.; Grand, R.S.; Allison, J.R.; Martienssen, R.A.; Nurse, P.; et al. A diffusion model for the coordination of DNA replication in Schizosaccharomyces pombe. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Ma, E.; Hyrien, O.; Goldar, A. Do replication forks control late origin firing in Saccharomyces cerevisiae? Nucleic Acids Res. 2012, 40, 2010–2019. [Google Scholar] [CrossRef]
- Shaw, A.; Olivares-Chauvet, P.; Maya-Mendoza, A.; Jackson, D.A. S-phase progression in mammalian cells: Modelling the influence of nuclear organization. Chromosome Res. 2010, 18, 163–178. [Google Scholar] [CrossRef]
- Guilbaud, G.; Rappailles, A.; Baker, A.; Chen, C.-L.; Arneodo, A.; Goldar, A.; d’Aubenton-Carafa, Y.; Thermes, C.; Audit, B.; Hyrien, O. Evidence for Sequential and Increasing Activation of Replication Origins along Replication Timing Gradients in the Human Genome. PLoS Comput. Biol. 2011, 7, e1002322. [Google Scholar] [CrossRef] [PubMed]
- Löb, D.; Lengert, N.; Chagin, V.O.; Reinhart, M.; Casas-Delucchi, C.S.; Cardoso, M.C.; Drossel, B. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun. 2016, 7, 11207. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Ge, X.Q. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009, 10, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.G.; Herrick, J.; Bechhoefer, J. Defects and DNA Replication. Phys. Rev. Lett. 2010, 104, 218104. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciardo, D.; Goldar, A.; Marheineke, K. On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway. Genes 2019, 10, 94. https://doi.org/10.3390/genes10020094
Ciardo D, Goldar A, Marheineke K. On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway. Genes. 2019; 10(2):94. https://doi.org/10.3390/genes10020094
Chicago/Turabian StyleCiardo, Diletta, Arach Goldar, and Kathrin Marheineke. 2019. "On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway" Genes 10, no. 2: 94. https://doi.org/10.3390/genes10020094




