Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions
Abstract
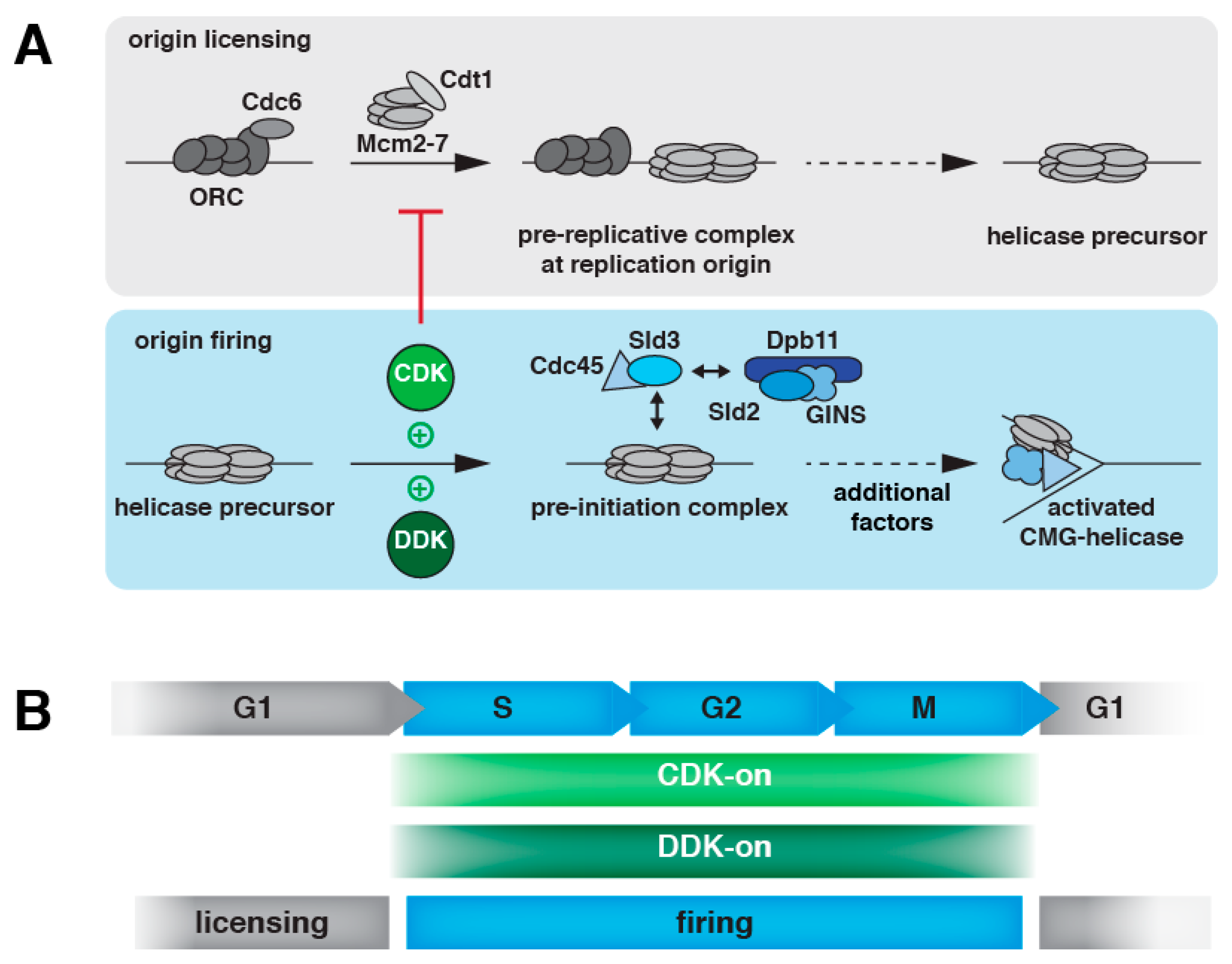
:1. Introduction
2. DNA Replication Initiation in Eukaryotes
2.1. DNA Replication Initiation Control in Budding Yeast
2.2. Additional DNA Replication Initiation Control Mechanisms in Metazoa
2.3. Deregulation of DNA Replication Initiation—Over-Replication and Genome Instability
2.4. Partial Deregulation of DNA Replication Initiation—Sporadic Over-Replication
3. DNA Replication Control at Cell Cycle Transitions
3.1. Bistable Switches—The Fundament of DNA Replication Control at Cell Cycle Transitions
3.2. Temporal Order of Licensing/Firing Activation/Inactivation at Cell Cycle Transitions
3.3. Intrinsic Temporal Order by CDK–Substrate Interactions
3.4. Temporal Order by Phosphatase–Substrate Interactions
3.5. Temporal Order by Degradation
3.6. Temporal Order by a Two-Kinase System
4. Conclusions and Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.; On, K.F.; Diffley, J.F.X. Regulating DNA Replication in Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a012930. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arias, E.E.; Walter, J.C. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007, 21, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.Q.; Co, C.; Li, J.J. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 2001, 411, 1068–1073. [Google Scholar] [CrossRef]
- Green, B.M.; Finn, K.J.; Li, J.J. Loss of DNA Replication Control Is a Potent Inducer of Gene Amplification. Science 2010, 329, 943–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, J.L.; Barrasa, M.I.; Orr-Weaver, T.L. Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr. Biol. 2015, 25, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Green, B.M.; Li, J.J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 2005, 16, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Kotsantis, P.; Petermann, E.; Boulton, S.J. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov. 2018, 8, 537–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hills, S.A.; Diffley, J.F.X. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2002, 2, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, K.M.; Hudson, T.J.; McPherson, J.D. Unraveling the genetics of cancer: Genome sequencing and beyond. Annu. Rev. Genom. Hum. Genet. 2011, 12, 407–430. [Google Scholar] [CrossRef] [PubMed]
- Bleichert, F.; Botchan, M.R.; Berger, J.M. Mechanisms for initiating cellular DNA replication. Science 2017, 355, eaah6317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507–516. [Google Scholar] [CrossRef]
- Gambus, A. Termination of Eukaryotic Replication Forks. Adv. Exp. Med. Biol. 2017, 1042, 163–187. [Google Scholar]
- Bhat, K.P.; Cortez, D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018, 25, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Szakal, B. Building up and breaking down: Mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F.X. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef] [Green Version]
- Donovan, S.; Harwood, J.; Drury, L.S.; Diffley, J.F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 5611–5616. [Google Scholar] [CrossRef] [Green Version]
- Seki, T.; Diffley, J.F. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 14115–14120. [Google Scholar] [CrossRef] [Green Version]
- Rowles, A.; Tada, S.; Blow, J.J. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999, 112 Pt 12, 2011–2018. [Google Scholar]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic Origin-Dependent DNA Replication In Vitro Reveals Sequential Action of DDK and S-CDK Kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Ilves, I.; Tamberg, N.; Petojevic, T.; Nogales, E.; Botchan, M.R.; Berger, J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011, 18, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aparicio, T.; Guillou, E.; Coloma, J.; Montoya, G.; Mendez, J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009, 37, 2087–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.P.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F.X. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurat, C.F.; Yeeles, J.T.P.; Patel, H.; Early, A.; Diffley, J.F.X. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol. Cell 2017, 65, 117–130. [Google Scholar] [CrossRef]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Azmi, I.F.; Watanabe, S.; Maloney, M.F.; Kang, S.; Belsky, J.A.; MacAlpine, D.M.; Peterson, C.L.; Bell, S.P. Nucleosomes influence multiple steps during replication initiation. eLife 2017, 6, e22512. [Google Scholar] [CrossRef]
- Coster, G.; Diffley, J.F.X. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 2017, 357, 314–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.; Diffley, J.F.X. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002, 4, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.F.; Beach, D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: Requirement for DNA replication and inhibition of mitosis. EMBO J. 1994, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Stillman, B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 1992, 357, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Weinreich, M.; Stillman, B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 1995, 81, 667–676. [Google Scholar] [CrossRef]
- Bueno, A.; Russell, P. Dual functions of CDC6: A yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992, 11, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef]
- Masumoto, H.; Sugino, A.; Araki, H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 2000, 20, 2809–2817. [Google Scholar] [CrossRef]
- Masumoto, H.; Muramatsu, S.; Kamimura, Y.; Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 2002, 415, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Diffley, J.F.X. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the MCM Complex. J. Biol. Chem. 2016, 291, 5879–5888. [Google Scholar] [CrossRef] [PubMed]
- Homesley, L.; Lei, M.; Kawasaki, Y.; Sawyer, S.; Christensen, T.; Tye, B.K. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000, 14, 913–926. [Google Scholar] [PubMed]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa-Onami, M.; Araki, H.; Tanaka, S. Pre-initiation complex assembly functions as a molecular switch that splits the Mcm2-7 double hexamer. EMBO Rep. 2017, 18, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; van Deursen, F.; De Piccoli, G.; Labib, K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013, 23, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Aves, S.J.; Liu, Y.; Richards, T.A. Evolutionary diversification of eukaryotic DNA replication machinery. Subcell. Biochem. 2012, 62, 19–35. [Google Scholar] [PubMed]
- Prioleau, M.-N.; MacAlpine, D.M. DNA replication origins-where do we begin? Genes Dev. 2016, 30, 1683–1697. [Google Scholar] [CrossRef]
- Gros, J.; Kumar, C.; Lynch, G.; Yadav, T.; Whitehouse, I.; Remus, D. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2-7 Sliding along DNA. Mol. Cell 2015, 60, 797–807. [Google Scholar] [CrossRef]
- Santocanale, C.; Diffley, J.F. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6671–6679. [Google Scholar] [CrossRef]
- Diffley, J.F.; Cocker, J.H.; Dowell, S.J.; Rowley, A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 1994, 78, 303–316. [Google Scholar] [CrossRef]
- Dahmann, C.; Diffley, J.F.; Nasmyth, K.A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995, 5, 1257–1269. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Collyer, T.; Hardy, C.F. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 1999, 19, 4270–4278. [Google Scholar] [CrossRef]
- Oshiro, G.; Owens, J.C.; Shellman, Y.; Sclafani, R.A.; Li, J.J. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol. 1999, 19, 4888–4896. [Google Scholar] [CrossRef] [PubMed]
- Pasero, P.; Duncker, B.P.; Schwob, E.; Gasser, S.M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 1999, 13, 2159–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, M.F.; Santocanale, C.; Drury, L.S.; Diffley, J.F. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 2000, 20, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Surana, U.; Robitsch, H.; Price, C.; Schuster, T.; Fitch, I.; Futcher, A.B.; Nasmyth, K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 1991, 65, 145–161. [Google Scholar] [CrossRef]
- Hadwiger, J.A.; Wittenberg, C.; Richardson, H.E.; de Barros Lopes, M.; Reed, S.I. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 1989, 86, 6255–6259. [Google Scholar] [CrossRef]
- Fitch, I.; Dahmann, C.; Surana, U.; Amon, A.; Nasmyth, K.; Goetsch, L.; Byers, B.; Futcher, B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1992, 3, 805–818. [Google Scholar] [CrossRef]
- Epstein, C.B.; Cross, F.R. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992, 6, 1695–1706. [Google Scholar] [CrossRef]
- Schwob, E.; Nasmyth, K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993, 7, 1160–1175. [Google Scholar] [CrossRef] [PubMed]
- Kitada, K.; Johnston, L.H.; Sugino, T.; Sugino, A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics 1992, 131, 21–29. [Google Scholar] [PubMed]
- Bishop, A.C.; Ubersax, J.A.; Petsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 407, 395–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, A.D.; Fangman, W.L.; Brewer, B.J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998, 12, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Woodbury, E.L.; Quang, P.N.; Paraz, M.; Blethrow, J.D.; Shah, K.; Shokat, K.M.; Morgan, D.O. Targets of the cyclin-dependent kinase Cdk1. Nature 2003, 425, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.J.; Tuch, B.B.; Villén, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef]
- Randell, J.C.W.; Fan, A.; Chan, C.; Francis, L.I.; Heller, R.C.; Galani, K.; Bell, S.P. Mec1 Is One of Multiple Kinases that Prime the Mcm2-7 Helicase for Phosphorylation by Cdc7. Mol. Cell 2010, 40, 353–363. [Google Scholar] [CrossRef]
- Sheu, Y.-J.; Stillman, B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef]
- Devault, A.; Gueydon, E.; Schwob, E. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol. Biol. Cell 2008, 19, 2267–2277. [Google Scholar] [CrossRef]
- Cho, W.-H.; Lee, Y.-J.; Kong, S.-I.; Hurwitz, J.; Lee, J.-K. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc. Natl. Acad. Sci. USA 2006, 103, 11521–11526. [Google Scholar] [CrossRef] [Green Version]
- Hardy, C.F.; Dryga, O.; Seematter, S.; Pahl, P.M.; Sclafani, R.A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA 1997, 94, 3151–3155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheu, Y.-J.; Stillman, B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010, 463, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Pahl, P.M.; Harrison, K.; Rosamond, J.; Sclafani, R.A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 1993, 13, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, S.; Hirai, K.; Tak, Y.-S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol epsilon, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.-S.; Tanaka, Y.; Endo, S.; Kamimura, Y.; Araki, H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006, 25, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Leem, S.H.; Phongdara, A.; Sugino, A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 1995, 92, 11791–11795. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Elledge, S.J. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 2002, 160, 1295–1304. [Google Scholar]
- Garcia, V.; Furuya, K.; Carr, A.M. Identification and functional analysis of TopBP1 and its homologs. DNA Repair 2005, 4, 1227–1239. [Google Scholar] [CrossRef]
- Puddu, F.; Granata, M.; Di Nola, L.; Balestrini, A.; Piergiovanni, G.; Lazzaro, F.; Giannattasio, M.; Plevani, P.; Muzi-Falconi, M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol. Cell. Biol. 2008, 28, 4782–4793. [Google Scholar] [CrossRef]
- Pfander, B.; Diffley, J.F.X. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011, 30, 4897–4907. [Google Scholar] [CrossRef] [Green Version]
- Gritenaite, D.; Princz, L.N.; Szakal, B.; Bantele, S.C.S.; Wendeler, L.; Schilbach, S.; Habermann, B.H.; Matos, J.; Lisby, M.; Branzei, D.; et al. A cell cycle-regulated Slx4-Dpb11 complex promotes the resolution of DNA repair intermediates linked to stalled replication. Genes Dev. 2014, 28, 1604–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bantele, S.C.; Ferreira, P.; Gritenaite, D.; Boos, D.; Pfander, B. Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. eLife 2017, 6, e21687. [Google Scholar] [CrossRef] [PubMed]
- Ohouo, P.Y.; de Oliveira, F.M.B.; Liu, Y.; Ma, C.J.; Smolka, M.B. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2012, 493, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, Y.; Tak, Y.S.; Sugino, A.; Araki, H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001, 20, 2097–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanemaki, M.; Labib, K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006, 25, 1753–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.; Nakato, R.; Katou, Y.; Shirahige, K.; Araki, H. Origin Association of Sld3, Sld7, and Cdc45 Proteins Is a Key Step for Determination of Origin-Firing Timing. Curr. Biol. 2011, 21, 2055–2063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deegan, T.D.; Yeeles, J.T.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016, 35, 961–973. [Google Scholar] [CrossRef]
- Takayama, Y.; Kamimura, Y.; Okawa, M.; Muramatsu, S.; Sugino, A.; Araki, H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003, 17, 1153–1165. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Bell, S.P. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011, 25, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Drury, L.S.; Perkins, G.; Diffley, J.F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997, 16, 5966–5976. [Google Scholar] [CrossRef] [Green Version]
- Drury, L.S.; Perkins, G.; Diffley, J.F. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000, 10, 231–240. [Google Scholar] [CrossRef] [Green Version]
- Elsasser, S.; Chi, Y.; Yang, P.; Campbell, J.L. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell 1999, 10, 3263–3277. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Zhang, W.; Koepp, D.M. The Hect-domain E3 ligase Tom1 and the F-box protein Dia2 control Cdc6 degradation in G1. J. Biol. Chem. 2012. [Google Scholar] [CrossRef]
- Labib, K.; Diffley, J.F.; Kearsey, S.E. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1999, 1, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Makino, N.; Nagai, M.; Araki, H.; Ushimaru, T. CDK phosphorylation regulates Mcm3 degradation in budding yeast. Biochem. Biophys. Res. Commun. 2018, 506, 680–684. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.; Liang, C.; Chen, H.H.; Stillman, B. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 2001, 98, 11211–11217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmes, G.M.; Archambault, V.; Austin, R.J.; Jacobson, M.D.; Bell, S.P.; Cross, F.R. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004, 18, 981–991. [Google Scholar] [CrossRef]
- Mimura, S.; Seki, T.; Tanaka, S.; Diffley, J.F.X. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 2004, 431, 1118–1123. [Google Scholar] [CrossRef]
- Drury, L.S.; Diffley, J.F.X. Factors Affecting the Diversity of DNA Replication Licensing Control in Eukaryotes. Curr. Biol. 2009, 19, 530–535. [Google Scholar] [CrossRef] [Green Version]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010, 140, 349–359. [Google Scholar] [CrossRef]
- Boos, D.; Sanchez-Pulido, L.; Rappas, M.; Pearl, L.H.; Oliver, A.W.; Ponting, C.P.; Diffley, J.F.X. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Curr. Biol. 2011, 21, 1152–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strausfeld, U.P.; Howell, M.; Rempel, R.; Maller, J.L.; Hunt, T.; Blow, J.J. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 1994, 4, 876–883. [Google Scholar] [CrossRef]
- Walter, J.C. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 2000, 275, 39773–39778. [Google Scholar] [CrossRef] [PubMed]
- Jares, P.; Blow, J.J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000, 14, 1528–1540. [Google Scholar] [PubMed]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Tada, S.; Li, A.; Maiorano, D.; Méchali, M.; Blow, J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001, 3, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGarry, T.J.; Kirschner, M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Quinn, L.M.; Herr, A.; McGarry, T.J.; Richardson, H. The Drosophila Geminin homolog: Roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001, 15, 2741–2754. [Google Scholar] [CrossRef]
- Yanagi, K.-I.; Mizuno, T.; Tsuyama, T.; Tada, S.; Iida, Y.; Sugimoto, A.; Eki, T.; Enomoto, T.; Hanaoka, F. Caenorhabditis elegans geminin homologue participates in cell cycle regulation and germ line development. J. Biol. Chem. 2005, 280, 19689–19694. [Google Scholar] [CrossRef]
- Arias, E.E.; Walter, J.C. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005, 19, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Arias, E.E.; Chen, J.; Harper, J.W.; Walter, J.C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 2006, 23, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 2006, 8, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Senga, T.; Sivaprasad, U.; Zhu, W.; Park, J.H.; Arias, E.E.; Walter, J.C.; Dutta, A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 2006, 281, 6246–6252. [Google Scholar] [CrossRef] [PubMed]
- Higa, L.A.; Banks, D.; Wu, M.; Kobayashi, R.; Sun, H.; Zhang, H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006, 5, 1675–1680. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xiong, Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 2006, 281, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Ralph, E.; Boye, E.; Kearsey, S.E. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006, 7, 1134–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, J.L.; Orr-Weaver, T.L. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 2016, 30, 2241–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neelsen, K.J.; Zanini, I.M.Y.; Mijic, S.; Herrador, R.; Zellweger, R.; Ray Chaudhuri, A.; Creavin, K.D.; Blow, J.J.; Lopes, M. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013, 27, 2537–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, B.M.; Morreale, R.J.; Ozaydin, B.; Derisi, J.L.; Li, J.J. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell 2006, 17, 2401–2414. [Google Scholar] [CrossRef] [PubMed]
- Finn, K.J.; Li, J.J. Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination. PLoS Genet. 2013, 9, e1003192. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Ikui, A.E.; Drapkin, B.J.; Cross, F.R. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 2005, 25, 6707–6721. [Google Scholar] [CrossRef] [PubMed]
- Tanny, R.E.; MacAlpine, D.M.; Blitzblau, H.G.; Bell, S.P. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell 2006, 17, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, S.L.; Li, J.J. Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy. PLoS Genet. 2015, 11, e1005039. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Rothstein, R.; Lisby, M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 2014, 198, 795–835. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Blow, J.J. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005, 24, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Takisawa, H.; Kubota, Y. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 2005, 10, 63–73. [Google Scholar] [CrossRef]
- Maiorano, D.; Krasinska, L.; Lutzmann, M.; Mechali, M. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr. Biol. 2005, 15, 146–153. [Google Scholar] [CrossRef]
- Melixetian, M.; Ballabeni, A.; Masiero, L.; Gasparini, P.; Zamponi, R.; Bartek, J.; Lukas, J.; Helin, K. Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 2004, 165, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 2004, 279, 30807–30816. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, Y.; Dutta, A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004, 24, 7140–7150. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.J.; Dutta, A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007, 21, 184–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Dutta, A. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol. Cell. Biol. 2006, 26, 4601–4611. [Google Scholar] [CrossRef] [PubMed]
- Diffley, J.F.X. Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 3545–3553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reußwig, K.-U.; Zimmermann, F.; Galanti, L.; Pfander, B. Robust Replication Control Is Generated by Temporal Gaps between Licensing and Firing Phases and Depends on Degradation of Firing Factor Sld2. Cell Rep. 2016, 17, 556–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Araki, H. Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance. PLoS Genet. 2011, 7, e1002136. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J.; Novak, B. Regulation of the eukaryotic cell cycle: Molecular antagonism, hysteresis, and irreversible transitions. J. Biol. 2001, 210, 249–263. [Google Scholar] [CrossRef]
- Chen, K.C.; Calzone, L.; Csikasz-Nagy, A.; Cross, F.R.; Novak, B.; Tyson, J.J. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 2004, 15, 3841–3862. [Google Scholar] [CrossRef] [PubMed]
- Cross, F.R. Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell 2003, 4, 741–752. [Google Scholar] [CrossRef]
- Santos, S.D.M.; Ferrell, J.E. Systems biology: On the cell cycle and its switches. Nature 2008, 454, 288–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skotheim, J.M.; Di Talia, S.; Siggia, E.D.; Cross, F.R. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 2008, 454, 291–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwob, E.; Böhm, T.; Mendenhall, M.D.; Nasmyth, K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 1994, 79, 233–244. [Google Scholar] [CrossRef]
- Verma, R.; Annan, R.S.; Huddleston, M.J.; Carr, S.A.; Reynard, G.; Deshaies, R.J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 1997, 278, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Pomerening, J.R.; Kim, S.Y.; Ferrell, J.E. Systems-level dissection of the cell-cycle oscillator: Bypassing positive feedback produces damped oscillations. Cell 2005, 122, 565–578. [Google Scholar] [CrossRef]
- Cross, F.R.; Archambault, V.; Miller, M.; Klovstad, M. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 2002, 13, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef]
- Brümmer, A.; Salazar, C.; Zinzalla, V.; Alberghina, L.; Höfer, T. Mathematical Modelling of DNA Replication Reveals a Trade-off between Coherence of Origin Activation and Robustness against Rereplication. PLoS Comput. Biol. 2010, 6, e1000783. [Google Scholar] [CrossRef]
- Loog, M.; Morgan, D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 2005, 434, 104–108. [Google Scholar] [CrossRef]
- Zhai, Y.; Yung, P.Y.K.; Huo, L.; Liang, C. Cdc14p resets the competency of replication licensing by dephosphorylating multiple initiation proteins during mitotic exit in budding yeast. J. Cell Sci. 2010, 123, 3933–3943. [Google Scholar] [CrossRef] [Green Version]
- Bloom, J.; Cross, F.R. Novel role for Cdc14 sequestration: Cdc14 dephosphorylates factors that promote DNA replication. Mol. Cell. Biol. 2007, 27, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Powers, B.L.; Hall, M.C. Re-examining the role of Cdc14 phosphatase in reversal of Cdk phosphorylation during mitotic exit. J. Cell Sci. 2017, 130, 2673–2681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchoux, C.; Uhlmann, F. A Quantitative Model for Ordered Cdk Substrate Dephosphorylation during Mitotic Exit. Cell 2011, 147, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishitani, H.; Sugimoto, N.; Roukos, V.; Nakanishi, Y.; Saijo, M.; Obuse, C.; Tsurimoto, T.; Nakayama, K.I.; Nakayama, K.; Fujita, M.; et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006, 25, 1126–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phizicky, D.V.; Berchowitz, L.E.; Bell, S.P. Multiple kinases inhibit origin licensing and helicase activation to ensure reductive cell division during meiosis. eLife 2018, 7, e33309. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Wagener, C.; Marinoni, F.; Kramer, E.R.; Melixetian, M.; Lazzerini Denchi, E.; Gieffers, C.; Matteucci, C.; Peters, J.M.; Helin, K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000, 14, 2330–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailand, N.; Diffley, J.F.X. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005, 122, 915–926. [Google Scholar] [CrossRef]
- Petersen, B.O.; Lukas, J.; Sørensen, C.S.; Bartek, J.; Helin, K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999, 18, 396–410. [Google Scholar] [CrossRef]
- Jiang, W.; Wells, N.J.; Hunter, T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 1999, 96, 6193–6198. [Google Scholar] [CrossRef] [Green Version]
- Saha, P.; Chen, J.; Thome, K.C.; Lawlis, S.J.; Hou, Z.H.; Hendricks, M.; Parvin, J.D.; Dutta, A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 1998, 18, 2758–2767. [Google Scholar] [CrossRef]
- Walter, D.; Hoffmann, S.; Komseli, E.-S.; Rappsilber, J.; Gorgoulis, V.; Sorensen, C.S. SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication. Nat. Commun. 2016, 7, 10530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Hsiao, J.Y.; Davey, N.E.; Van Voorhis, V.A.; Foster, S.A.; Tang, C.; Morgan, D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014, 207, 23–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamenz, J.; Mihaljev, T.; Kubis, A.; Legewie, S.; Hauf, S. Robust Ordering of Anaphase Events by Adaptive Thresholds and Competing Degradation Pathways. Mol. Cell 2015, 60, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Coleman, K.E.; Grant, G.D.; Haggerty, R.A.; Brantley, K.; Shibata, E.; Workman, B.D.; Dutta, A.; Varma, D.; Purvis, J.E.; Cook, J.G. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 2015, 29, 1734–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantiero, D.; Mackenzie, A.; Donaldson, A.; Zegerman, P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011, 30, 4805–4814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zegerman, P.; Diffley, J.F.X. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 2010, 467, 474–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiraga, S.-I.; Alvino, G.M.; Chang, F.; Lian, H.-Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef] [Green Version]
- Weinreich, M.; Stillman, B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999, 18, 5334–5346. [Google Scholar] [CrossRef] [Green Version]
- Mattarocci, S.; Shyian, M.; Lemmens, L.; Damay, P.; Altintas, D.M.; Shi, T.; Bartholomew, C.R.; Thomä, N.H.; Hardy, C.F.J.; Shore, D. Rif1 Controls DNA Replication Timing in Yeast through the PP1 Phosphatase Glc7. Cell Rep. 2014, 7, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Davé, A.; Cooley, C.; Garg, M.; Bianchi, A. Protein Phosphatase 1 Recruitment by Rif1 Regulates DNA Replication Origin Firing by Counteracting DDK Activity. Cell Rep. 2014, 7, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Zhao, X. A new MCM modification cycle regulates DNA replication initiation. Nat. Struct. Mol. Biol. 2016, 23, 209–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nougarède, R.; Della Seta, F.; Zarzov, P.; Schwob, E. Hierarchy of S-phase-promoting factors: Yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 2000, 20, 3795–3806. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reusswig, K.-U.; Pfander, B. Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes 2019, 10, 99. https://doi.org/10.3390/genes10020099
Reusswig K-U, Pfander B. Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes. 2019; 10(2):99. https://doi.org/10.3390/genes10020099
Chicago/Turabian StyleReusswig, Karl-Uwe, and Boris Pfander. 2019. "Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions" Genes 10, no. 2: 99. https://doi.org/10.3390/genes10020099
APA StyleReusswig, K.-U., & Pfander, B. (2019). Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes, 10(2), 99. https://doi.org/10.3390/genes10020099




