Headcase is a Repressor of Lamellocyte Fate in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks
2.2. Antibodies
2.3. P Element Conversion
2.4. X-GAL Staining
2.5. PCR Mapping of the P Element Insertions
2.6. Generation of hdc Alleles and Breakpoint Mapping
2.7. Immunostaining of Circulating Hemocytes, Imaging, and Counting
2.8. Preparation and Immunostaining of the Lymph Gland
3. Results
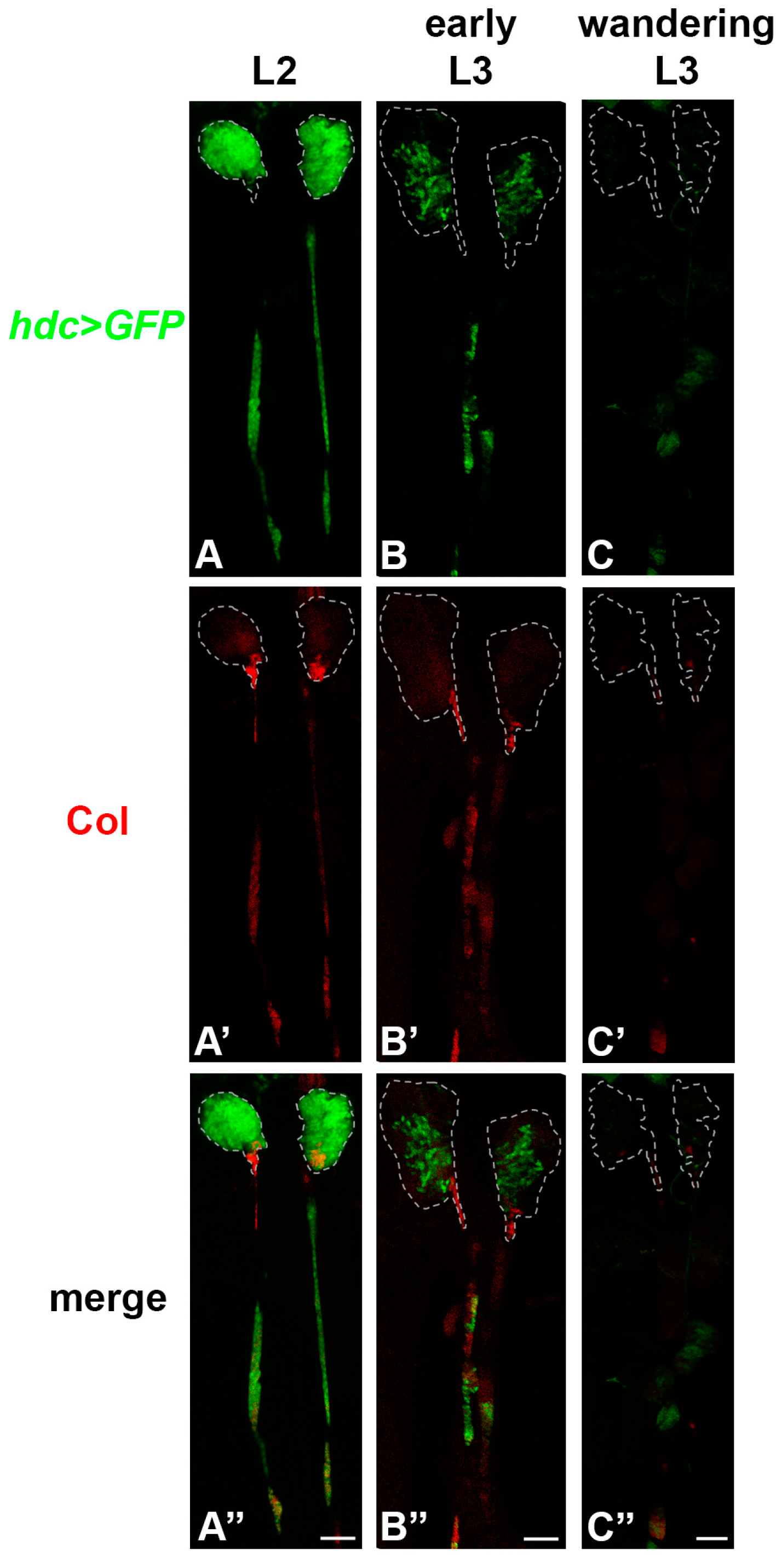
3.1. hdc is Expressed in the Lymph Gland, but Not in the Other Hemocyte Compartments
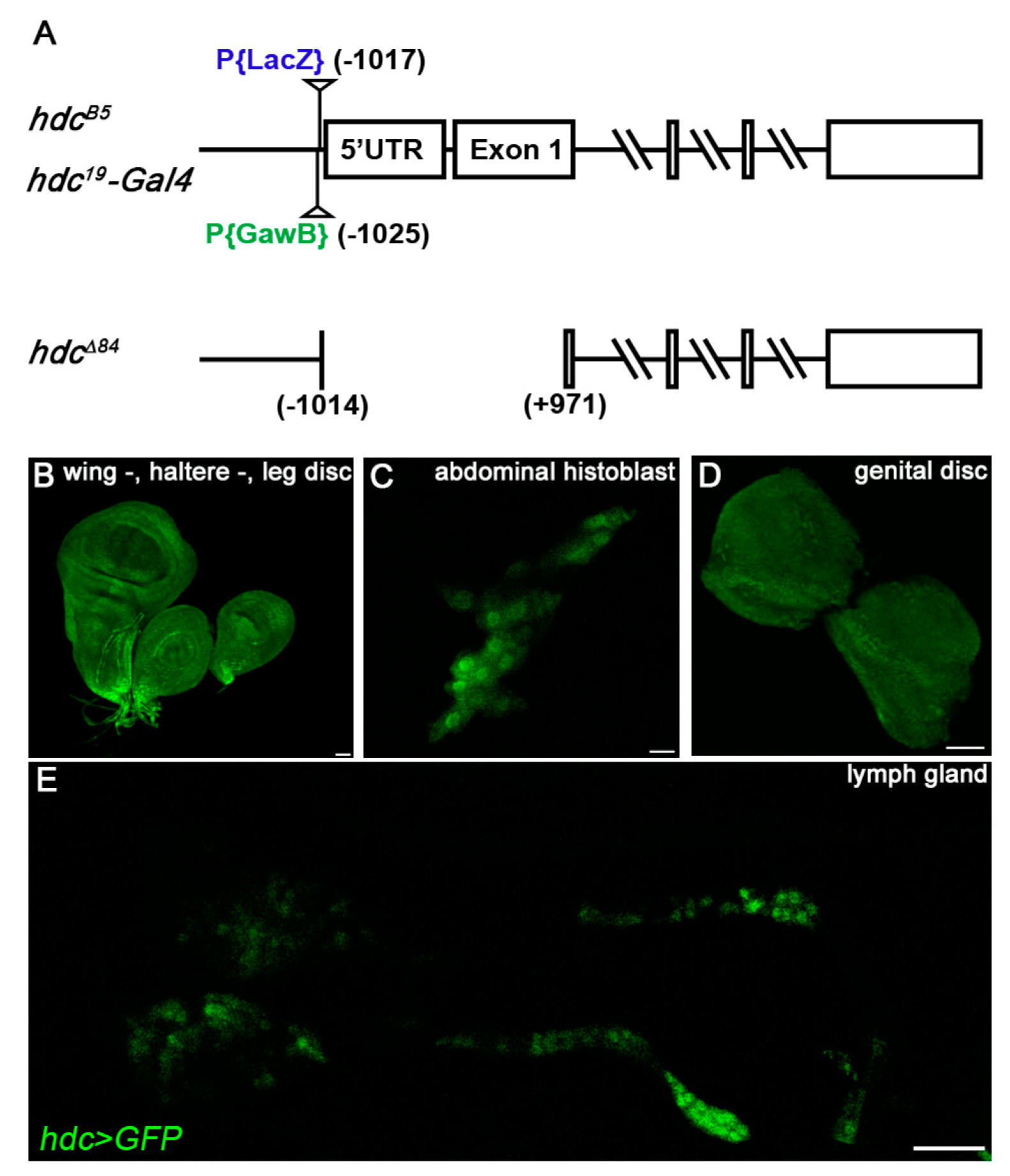
3.2. hdc Enhancer Trap Insertions are Hypomorphic hdc Alleles
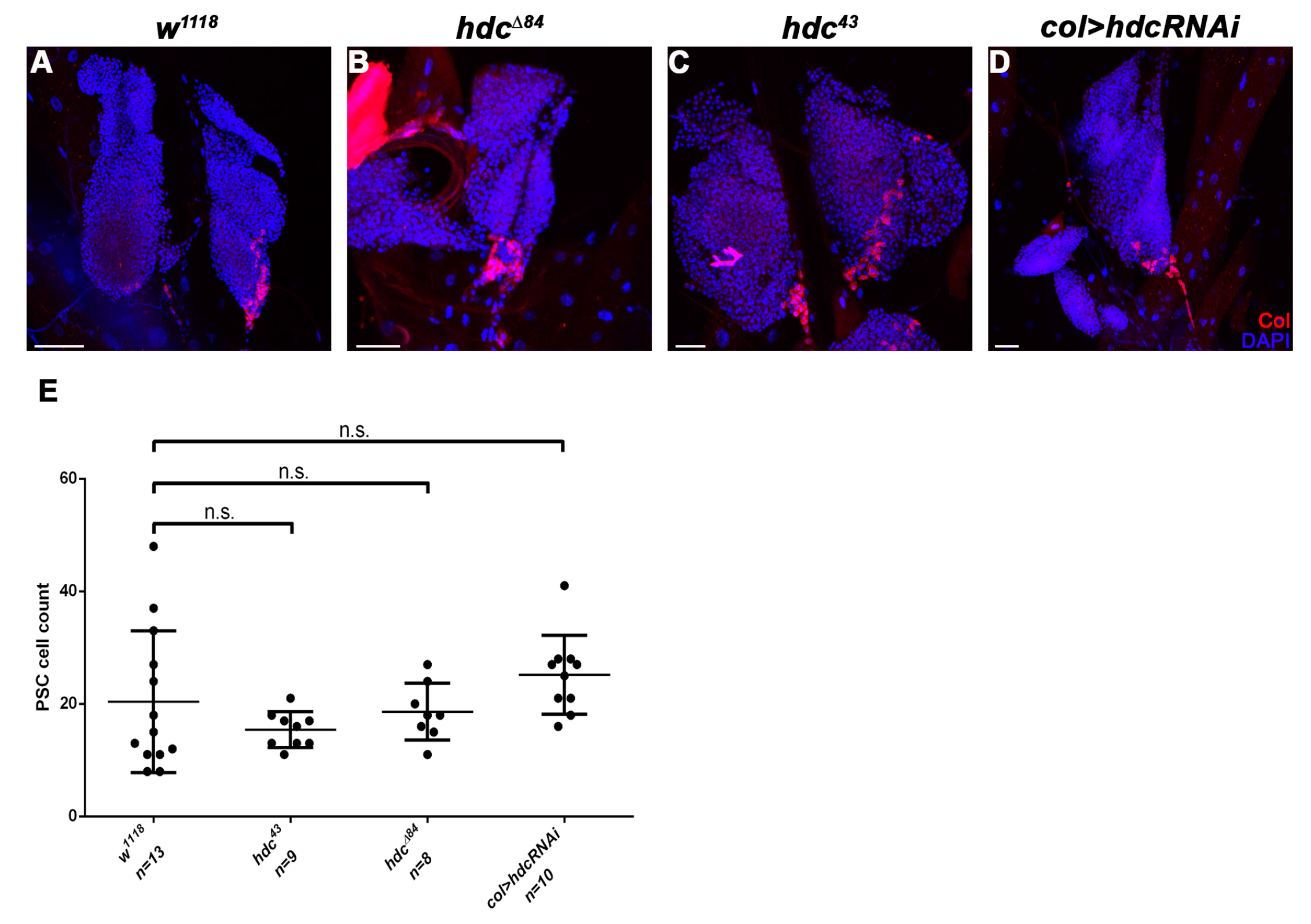
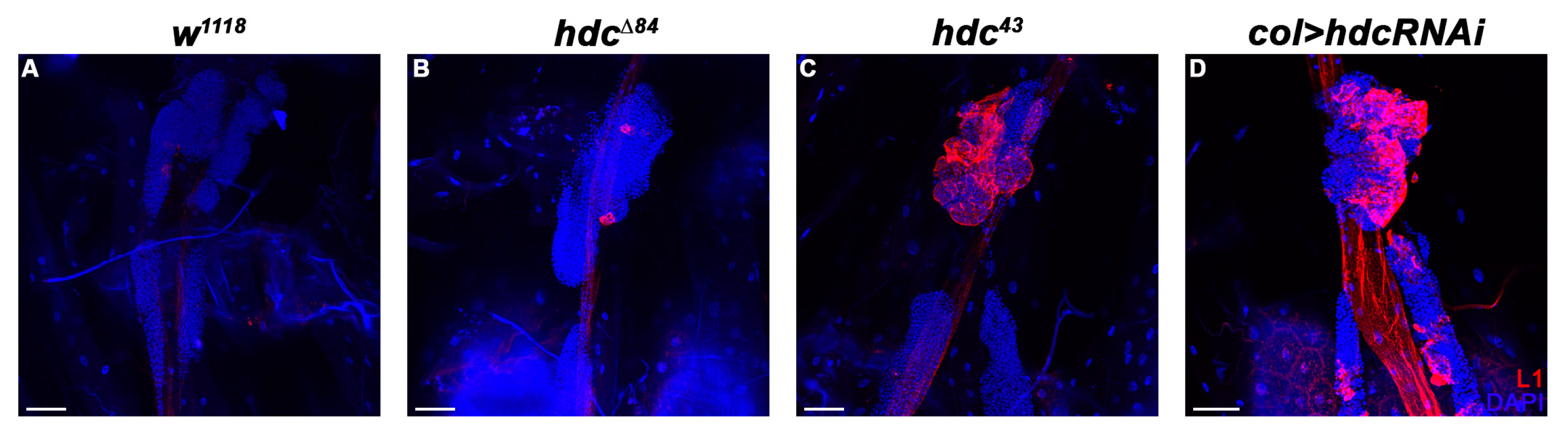
3.3. hdc Loss-Of-Function Mutant Larvae Produce Lamellocytes without Immune Induction
3.4. Hdc Exerts its Effect in the Lymph Gland
3.5. Hdc Interacts with the Hedgehog and the Decapentaplegic Pathways
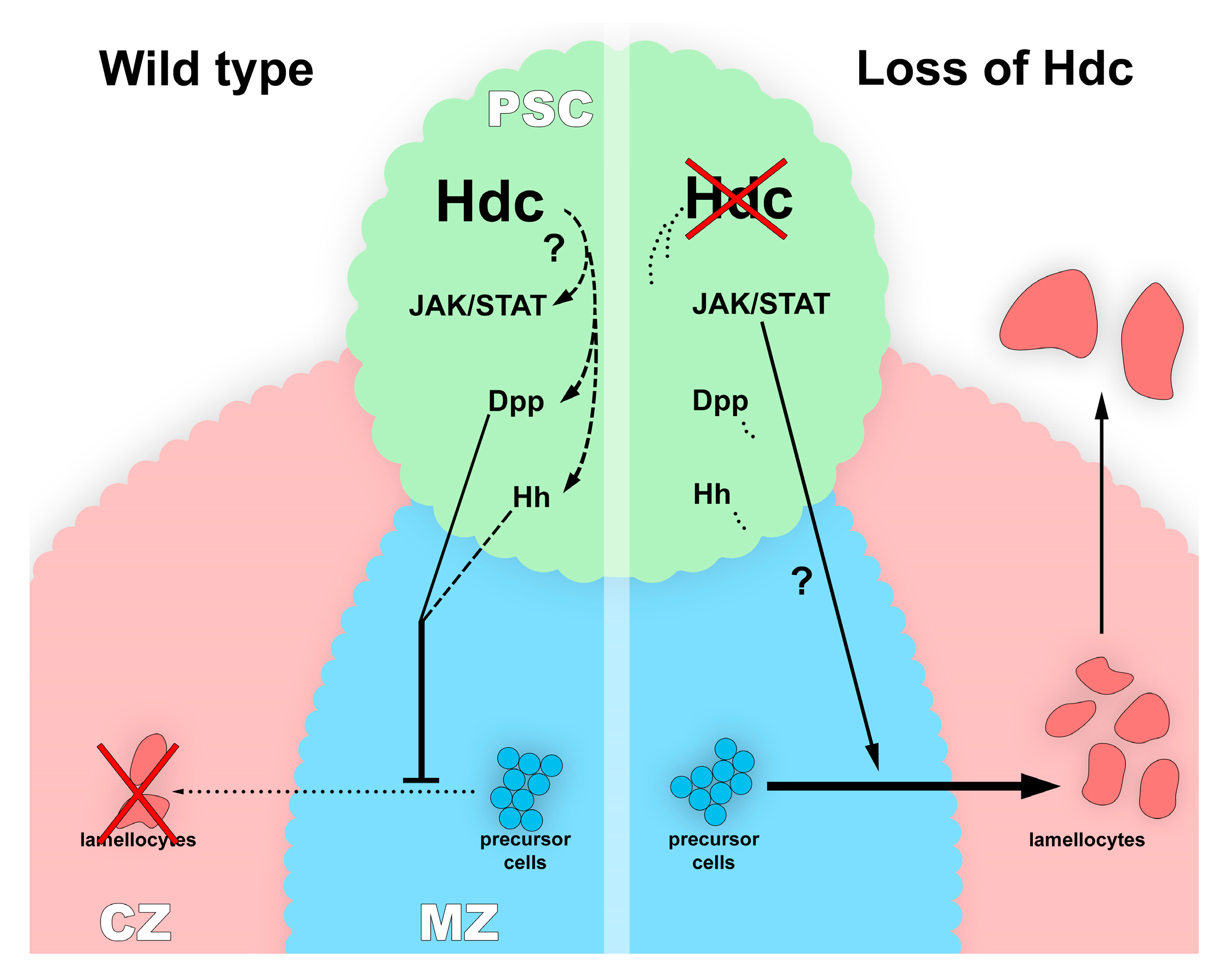
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Becker, A.J.; McCulloch, E.A.; Till, J.E. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 1963, 197, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.P.; Alexander, W.S. Haematopoietic stem cells: Past, present and future. Cell Death Discov. 2017, 3, 17002. [Google Scholar] [CrossRef] [PubMed]
- Boulais, P.E.; Frenette, P.S. Making sense of hematopoietic stem cell niches. Blood 2015, 125, 2621–2629. [Google Scholar] [CrossRef] [PubMed]
- Asada, N.; Takeishi, S.; Frenette, P.S. Complexity of bone marrow hematopoietic stem cell niche. Int. J. Hematol. 2017, 106, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Tamma, R.; Ribatti, D. Bone Niches, Hematopoietic Stem Cells, and Vessel Formation. Int. J. Mol. Sci. 2017, 18, 151. [Google Scholar] [CrossRef] [PubMed]
- Calvi, L.M.; Link, D.C. The hematopoietic stem cell niche in homeostasis and disease. Blood 2015, 126, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.-C.; Link, D.C. Concise Review: The Malignant Hematopoietic Stem Cell Niche. Stem Cells 2017, 35, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Hartenstein, V.; Banerjee, U. Thicker than blood: Conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 2003, 5, 673–690. [Google Scholar] [CrossRef]
- Honti, V.; Csordás, G.; Kurucz, É.; Márkus, R.; Andó, I. The cell-mediated immunity of Drosophila melanogaster: Hemocyte lineages, immune compartments, microanatomy and regulation. Dev. Comp. Immunol. 2014, 42, 47–56. [Google Scholar] [CrossRef]
- Gold, K.S.; Brückner, K. Macrophages and cellular immunity in Drosophila melanogaster. Semin. Immunol. 2015, 27, 357–368. [Google Scholar] [CrossRef]
- Letourneau, M.; Lapraz, F.; Sharma, A.; Vanzo, N.; Waltzer, L.; Crozatier, M. Drosophila hematopoiesis under normal conditions and in response to immune stress. FEBS Lett. 2016, 590, 4034–4051. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Kadowaki, T.; Kitagawa, Y. Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev. Biol. 2003, 264, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, E.; Zettervall, C.-J.; Sinka, R.; Vilmos, P.; Pivarcsi, A.; Ekengren, S.; Hegedüs, Z.; Ando, I.; Hultmark, D. Hemese, a hemocyte-specific transmembrane protein, affects the cellular immune response in Drosophila. Proc. Natl. Acad. Sci. USA 2003, 100, 2622–2627. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, E.; Márkus, R.; Zsámboki, J.; Folkl-Medzihradszky, K.; Darula, Z.; Vilmos, P.; Udvardy, A.; Krausz, I.; Lukacsovich, T.; Gateff, E.; Zettervall, C.-J.; Hultmark, D.; Andó, I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 2007, 17, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, E.; Váczi, B.; Márkus, R.; Laurinyecz, B.; Vilmos, P.; Zsámboki, J.; Csorba, K.; Gateff, E.; Hultmark, D.; Andó, I. Definition of Drosophila hemocyte subsets by cell-type specific antigens. Acta. Biol. Hung. 2007, 58, 95–111. [Google Scholar] [CrossRef]
- Honti, V.; Kurucz, E.; Csordás, G.; Laurinyecz, B.; Márkus, R.; Andó, I. In vivo detection of lamellocytes in Drosophila melanogaster. Immunol. Lett. 2009, 126, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Tokusumi, T.; Shoue, D.A.; Tokusumi, Y.; Stoller, J.R.; Schulz, R.A. New hemocyte-specific enhancer-reporter transgenes for the analysis of hematopoiesis in Drosophila. Genesis 2009, 47, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Liu, T.; Banerjee, U. Drosophila hematopoiesis: Markers and methods for molecular genetic analysis. Methods 2014, 68, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.J.; Vass, E.; Frey, F.; Carton, Y. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur. J. Cell Biol. 1995, 68, 450–456. [Google Scholar]
- Russo, J.; Dupas, S.; Frey, F.; Carton, Y.; Brehelin, M. Insect immunity: Early events in the encapsulation process of parasitoid (Leptopilina boulardi) eggs in resistant and susceptible strains of Drosophila. Parasitology 1996, 112 Pt 1, 135–142. [Google Scholar] [CrossRef]
- Lanot, R.; Zachary, D.; Holder, F.; Meister, M. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 2001, 230, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Honti, V.; Csordás, G.; Márkus, R.; Kurucz, E.; Jankovics, F.; Andó, I. Cell lineage tracing reveals the plasticity of the hemocyte lineages and of the hematopoietic compartments in Drosophila melanogaster. Mol. Immunol. 2010, 47, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, R.P.; Carton, Y.; Govind, S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 2002, 243, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Avet-Rochex, A.; Boyer, K.; Polesello, C.; Gobert, V.; Osman, D.; Roch, F.; Augé, B.; Zanet, J.; Haenlin, M.; Waltzer, L. An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis. BMC Dev. Biol. 2010, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Krzemien, J.; Oyallon, J.; Crozatier, M.; Vincent, A. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev. Biol. 2010, 346, 310–319. [Google Scholar] [CrossRef]
- Stofanko, M.; Kwon, S.Y.; Badenhorst, P. Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity. PLoS ONE 2010, 5, e14051. [Google Scholar] [CrossRef] [PubMed]
- Anderl, I.; Vesala, L.; Ihalainen, T.O.; Vanha-Aho, L.-M.; Andó, I.; Rämet, M.; Hultmark, D. Transdifferentiation and Proliferation in Two Distinct Hemocyte Lineages in Drosophila melanogaster Larvae after Wasp Infection. PLoS Pathog. 2016, 12, e1005746. [Google Scholar] [CrossRef] [PubMed]
- Márkus, R.; Laurinyecz, B.; Kurucz, E.; Honti, V.; Bajusz, I.; Sipos, B.; Somogyi, K.; Kronhamn, J.; Hultmark, D.; Andó, I. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2009, 106, 4805–4809. [Google Scholar] [CrossRef]
- Kroeger, P.T.; Tokusumi, T.; Schulz, R.A. Transcriptional regulation of eater gene expression in Drosophila blood cells. Genesis 2012, 50, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Crozatier, M.; Ubeda, J.-M.; Vincent, A.; Meister, M. Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol. 2004, 2, E196. [Google Scholar] [CrossRef] [PubMed]
- Krzemień, J.; Dubois, L.; Makki, R.; Meister, M.; Vincent, A.; Crozatier, M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 2007, 446, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Mandal, L.; Martinez-Agosto, J.A.; Evans, C.J.; Hartenstein, V.; Banerjee, U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature 2007, 446, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Pennetier, D.; Oyallon, J.; Morin-Poulard, I.; Dejean, S.; Vincent, A.; Crozatier, M. Size control of the Drosophila hematopoietic niche by bone morphogenetic protein signaling reveals parallels with mammals. Proc. Natl. Acad. Sci. USA 2012, 109, 3389–3394. [Google Scholar] [CrossRef] [PubMed]
- Morin-Poulard, I.; Sharma, A.; Louradour, I.; Vanzo, N.; Vincent, A.; Crozatier, M. Vascular control of the Drosophila haematopoietic microenvironment by Slit/Robo signalling. Nat. Commun. 2016, 7, 11634. [Google Scholar] [CrossRef] [PubMed]
- Khadilkar, R.J.; Vogl, W.; Goodwin, K.; Tanentzapf, G. Modulation of occluding junctions alters the hematopoietic niche to trigger immune activation. Elife 2017, 6, e28081. [Google Scholar] [CrossRef] [PubMed]
- Benmimoun, B.; Polesello, C.; Haenlin, M.; Waltzer, L. The EBF transcription factor Collier directly promotes Drosophila blood cell progenitor maintenance independently of the niche. Proc. Natl. Acad. Sci. USA 2015, 112, 9052–9057. [Google Scholar] [CrossRef] [PubMed]
- Oyallon, J.; Vanzo, N.; Krzemień, J.; Morin-Poulard, I.; Vincent, A.; Crozatier, M. Two Independent Functions of Collier/Early B Cell Factor in the Control of Drosophila Blood Cell Homeostasis. PLoS ONE 2016, 11, e0148978. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-H.; Evans, C.J.; Uemura, C.; Banerjee, U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development 2005, 132, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Luo, F.; Jin, L.H. The Drosophila lymph gland is an ideal model for studying hematopoiesis. Dev. Comp. Immunol. 2017, 83, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Morin-Poulard, I.; Vincent, A.; Crozatier, M. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT 2013, 2, e25700. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.S.; Ramesh, P.; Chugh, M.; Mandal, S.; Mandal, L. Dpp dependent Hematopoietic stem cells give rise to Hh dependent blood progenitors in larval lymph gland of Drosophila. Elife 2016, 5, e18295. [Google Scholar] [CrossRef] [PubMed]
- Makki, R.; Meister, M.; Pennetier, D.; Ubeda, J.-M.; Braun, A.; Daburon, V.; Krzemień, J.; Bourbon, H.-M.; Zhou, R.; Vincent, A.; Crozatier, M. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010, 8, e1000441. [Google Scholar] [CrossRef] [PubMed]
- Fossett, N. Signal transduction pathways, intrinsic regulators, and the control of cell fate choice. Biochim. Biophys. Acta 2013, 1830, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Baldeosingh, R.; Gao, H.; Wu, X.; Fossett, N. Hedgehog signaling from the Posterior Signaling Center maintains U-shaped expression and a prohemocyte population in Drosophila. Dev. Biol. 2018, 441, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Tan, W.; Steward, R. JAK/STAT and the GATA factor Pannier control hemocyte maturation and differentiation in Drosophila. Dev. Biol. 2011, 352, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Grigorian, M.; Mandal, L.; Hartenstein, V. Hematopoiesis at the onset of metamorphosis: Terminal differentiation and dissociation of the Drosophila lymph gland. Dev. Genes Evol. 2011, 221, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, A.J.; Honti, V.; Binggeli, O.; Burri, O.; Poidevin, M.; Kurucz, É.; Zsámboki, J.; Andó, I.; Lemaitre, B. The Nimrod transmembrane receptor Eater is required for hemocyte attachment to the sessile compartment in Drosophila melanogaster. Biol. Open 2015, 4, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-C.; Chang, C.-C.; Yang, S.-H.; Chen, S.-H.; Huang, C.-J. A homologue of the Drosophila headcase protein is a novel tumor marker for early-stage colorectal cancer. Oncol. Rep. 2006, 15, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Dowejko, A.; Bauer, R.J.; Müller-Richter, U.D.A.; Reichert, T.E. The human homolog of the Drosophila headcase protein slows down cell division of head and neck cancer cells. Carcinogenesis 2009, 30, 1678–1685. [Google Scholar] [CrossRef] [PubMed]
- Dowejko, A.; Bauer, R.; Bauer, K.; Müller-Richter, U.D.A.; Reichert, T.E. The human HECA interacts with cyclins and CDKs to antagonize Wnt-mediated proliferation and chemoresistance of head and neck cancer cells. Exp. Cell Res. 2012, 318, 489–499. [Google Scholar] [CrossRef]
- Wang, J.; Gong, L.; Zhu, S.-J.; Zhu, Q.; Yao, L.; Han, X.-J.; Zhang, J.-R.; Li, Y.-H.; Zhang, W. The Human Homolog of Drosophila Headcase Acts as a Tumor Suppressor through Its Blocking Effect on the Cell Cycle in Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0137579. [Google Scholar] [CrossRef] [PubMed]
- Weaver, T.A.; White, R.A. headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development 1995, 121, 4149–4160. [Google Scholar] [PubMed]
- Resende, L.P.F.; Boyle, M.; Tran, D.; Fellner, T.; Jones, D.L. Headcase promotes cell survival and niche maintenance in the Drosophila testis. PLoS ONE 2013, 8, e68026. [Google Scholar] [CrossRef] [PubMed]
- Resende, L.P.F.; Truong, M.E.; Gomez, A.; Jones, D.L. Intestinal stem cell ablation reveals differential requirements for survival in response to chemical challenge. Dev. Biol. 2017, 424, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Sinenko, S.A.; Mathey-Prevot, B. Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene 2004, 23, 9120–9128. [Google Scholar] [CrossRef] [PubMed]
- Sepp, K.J.; Auld, V.J. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics 1999, 151, 1093–1101. [Google Scholar] [PubMed]
- Jankovics, F.; Sinka, R.; Erdélyi, M. An Interaction Type of Genetic Screen Reveals a Role of the Rab11 Gene in oskar mRNA Localization in the Developing Drosophila melanogaster Oocyte. Genetics 2001, 158, 1177–1188. [Google Scholar]
- Kimbrell, D.A.; Hice, C.; Bolduc, C.; Kleinhesselink, K.; Beckingham, K. The Dorothy enhancer has Tinman binding sites and drives hopscotch-induced tumor formation. Genesis 2002, 34, 23–28. [Google Scholar] [CrossRef]
- Evans, C.J.; Olson, J.M.; Ngo, K.T.; Kim, E.; Lee, N.E.; Kuoy, E.; Patananan, A.N.; Sitz, D.; Tran, P.; Do, M.-T.; Yackle, K.; Cespedes, A.; Hartenstein, V.; Call, G.B.; Banerjee, U. G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 2009, 6, 603–605. [Google Scholar] [CrossRef]
- Binari, R.; Perrimon, N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994, 8, 300–312. [Google Scholar] [CrossRef]
- Ríos-Barrera, L.D.; Riesgo-Escovar, J.R. Regulating cell morphogenesis: The Drosophila Jun N-terminal kinase pathway. Genesis 2013, 51, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.R.; Anderl, I.; Vo, H.T.M.; Valanne, S.; Yang, H.; Kronhamn, J.; Rämet, M.; Rusten, T.E.; Hultmark, D. Genetic Screen in Drosophila Larvae Links ird1 Function to Toll Signaling in the Fat Body and Hemocyte Motility. PLoS ONE 2016, 11, e0159473. [Google Scholar] [CrossRef] [PubMed]
- Giordani, G.; Barraco, M.; Giangrande, A.; Martinelli, G.; Guadagnuolo, V.; Simonetti, G.; Perini, G.; Bernardoni, R. The human Smoothened inhibitor PF-04449913 induces exit from quiescence and loss of multipotent Drosophila hematopoietic progenitor cells. Oncotarget 2016, 7, 55313–55327. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, X.; Fossett, N. Upregulation of the Drosophila Friend of GATA gene U-shaped by JAK/STAT signaling maintains lymph gland prohemocyte potency. Mol. Cell. Biol. 2009, 29, 6086–6096. [Google Scholar] [CrossRef] [PubMed]
- Amoyel, M.; Bach, E. Functions of the Drosophila JAK-STAT pathway. JAKSTAT 2012, 1, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Sinenko, S.A.; Shim, J.; Banerjee, U. Oxidative stress in the haematopoietic niche regulates the cellular immune response in Drosophila. EMBO Rep. 2011, 13, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Louradour, I.; Sharma, A.; Morin-Poulard, I.; Letourneau, M.; Vincent, A.; Crozatier, M.; Vanzo, N. Reactive oxygen species-dependent Toll/NF-κB activation in the Drosophila hematopoietic niche confers resistance to wasp parasitism. Elife 2017, 6, e25496. [Google Scholar] [CrossRef]
- Steneberg, P.; Englund, C.; Kronhamn, J.; Weaver, T.A.; Samakovlis, C. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev. 1998, 12, 956–967. [Google Scholar] [CrossRef]
- Estella, C.; Rieckhof, G.; Calleja, M.; Morata, G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development 2003, 130, 5929–5941. [Google Scholar] [CrossRef]
- Avet-Rochex, A.; Carvajal, N.; Christoforou, C.P.; Yeung, K.; Maierbrugger, K.T.; Hobbs, C.; Lalli, G.; Cagin, U.; Plachot, C.; McNeill, H.; Bateman, J.M. Unkempt is negatively regulated by mTOR and uncouples neuronal differentiation from growth control. PLoS Genet. 2014, 10, e1004624. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varga, G.I.B.; Csordás, G.; Cinege, G.; Jankovics, F.; Sinka, R.; Kurucz, É.; Andó, I.; Honti, V. Headcase is a Repressor of Lamellocyte Fate in Drosophila melanogaster. Genes 2019, 10, 173. https://doi.org/10.3390/genes10030173
Varga GIB, Csordás G, Cinege G, Jankovics F, Sinka R, Kurucz É, Andó I, Honti V. Headcase is a Repressor of Lamellocyte Fate in Drosophila melanogaster. Genes. 2019; 10(3):173. https://doi.org/10.3390/genes10030173
Chicago/Turabian StyleVarga, Gergely I. B., Gábor Csordás, Gyöngyi Cinege, Ferenc Jankovics, Rita Sinka, Éva Kurucz, István Andó, and Viktor Honti. 2019. "Headcase is a Repressor of Lamellocyte Fate in Drosophila melanogaster" Genes 10, no. 3: 173. https://doi.org/10.3390/genes10030173





