Molecular Evolution of Tryptophan Hydroxylases in Vertebrates: A Comparative Genomic Survey
Abstract
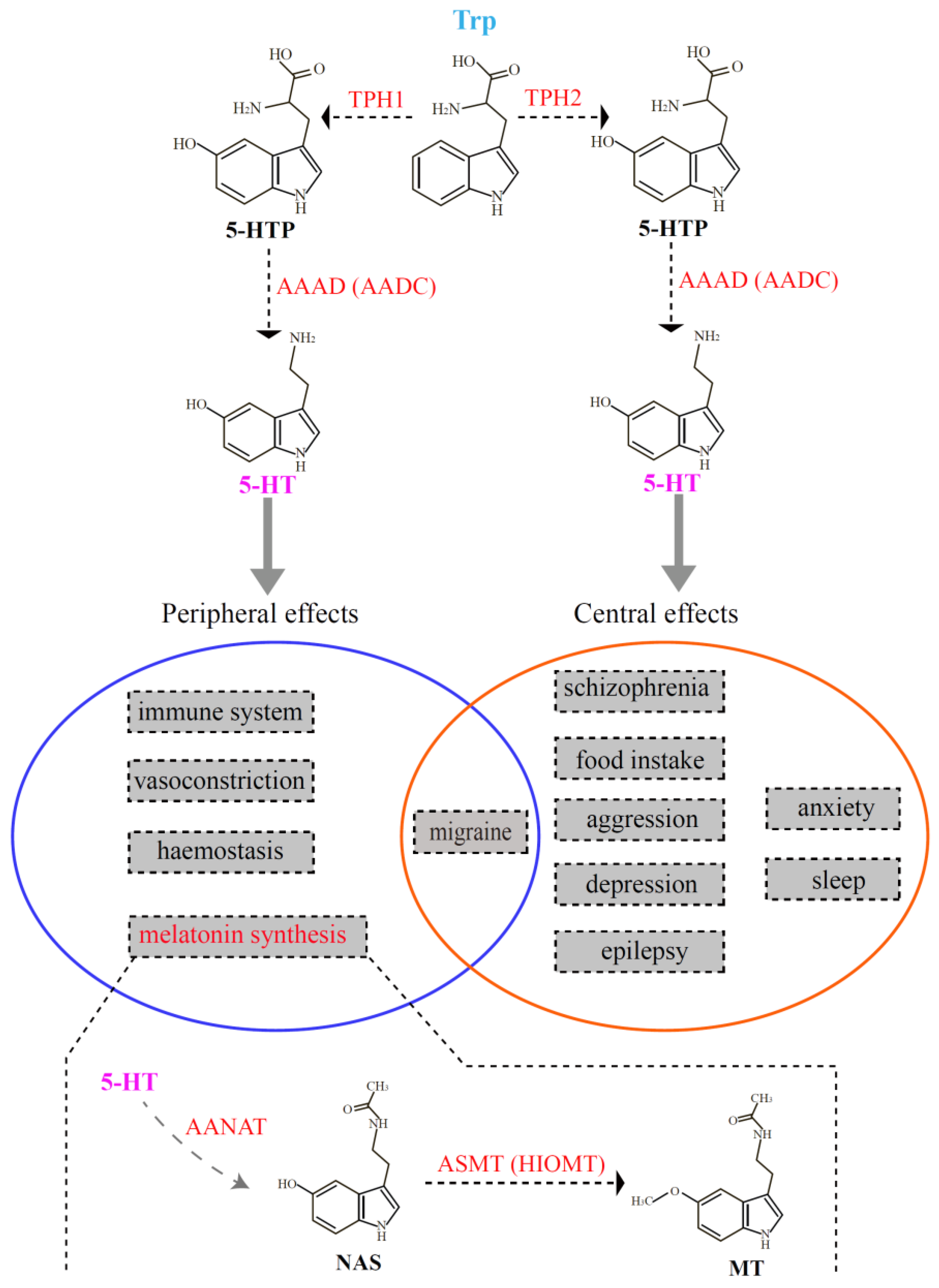
:1. Introduction
2. Materials and Methods
2.1. Sequence Collection
2.2. Sequence Alignment and Phylogenetic Reconstruction
2.3. Identification of Conserved Synteny
2.4. Detection of Differences between TPH Protein Sequences
2.5. Prediction of Tertiary Structures of TPH Proteins
2.6. Identification of Putative Positively Selected Sites
3. Results
3.1. Variation of tph Copy Number in Vertebrates
3.2. Phylogenetic Relationships among the tph Genes in Vertebrates
3.3. Synteny Data
3.4. Sequence Variations and Secondary Structures of the tph Genes
3.5. Predicted Three-Dimensional (3D) Structures of the TPH Proteins
3.6. Detection of Putative Positively Selected Sites in the TPH Family
4. Discussion
4.1. Possible Reasons for Variation of tph Copy Number in Vertebrates
4.2. Adaptive Evolution of TPHs in Vertebrates and Impact on Human Health
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Lucki, I. The spectrum of behaviors influenced by serotonin. Biol. Psychiatry 1998, 44, 151–162. [Google Scholar] [CrossRef]
- Jacobs, B.L.; Azmitia, E.C. Structure and function of the brain serotonin system. Physiol. Rev. 1992, 72, 165–229. [Google Scholar] [CrossRef] [PubMed]
- Côté, F.; Fligny, C.; Bayard, E.; Launay, J.-M.; Gershon, M.D.; Mallet, J.; Vodjdani, G. Maternal serotonin is crucial for murine embryonic development. Proc. Natl. Acad. Sci. USA 2007, 104, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Chong, N.W.; Bernard, M.; Klein, D.C. Characterization of the chicken serotonin N-acetyltransferase gene. J. Biol. Chem. 2000, 275, 32991–32998. [Google Scholar] [CrossRef] [PubMed]
- Lovenberg, W.; Weissbach, H.; Udenfriend, S. Aromatic l-amino acid decarboxylase. J. Biol. Chem. 1962, 237, 89. [Google Scholar] [PubMed]
- Bernard, M.; Iuvone, P.M.; Cassone, V.M.; Roseboom, P.H.; Coon, S.L.; Klein, D.C. Avian melatonin synthesis: Photic and circadian regulation of serotonin N-acetyltransferase mRNA in the chicken pineal gland and retina. J. Neurochem. 1997, 68, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Ishida, I.; Obinata, M.; Deguchi, T. Molecular cloning and nucleotide sequence of cDNA encoding hydroxyindole o-methyltransferase of bovine pineal glands. J. Biol. Chem. 1987, 262, 2895–2899. [Google Scholar]
- McKinney, J.; Teigen, K.; Frøystein, N.Å.; Salaün, C.; Knappskog, P.M.; Haavik, J.; Martínez, A. Conformation of the substrate and pterin cofactor bound to human tryptophan hydroxylase. Important role of Phe313 in substrate specificity. Biochemistry 2001, 40, 15591–15601. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.F. Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 1999, 68, 355. [Google Scholar] [CrossRef]
- Erlandsen, H.; Bjørgo, E.; Flatmark, T.; Stevens, R.C. Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase. Biochemistry 2000, 39, 2208–2217. [Google Scholar] [CrossRef] [PubMed]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Andersen, O.A.; Flatmark, T.; Hough, E. Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-l-alanine, and its implications for the mechanism of catalysis and substrate activation. J. Mol. Biol. 2002, 320, 1095–1108. [Google Scholar] [CrossRef]
- Walther, D.J.; Peter, J.U.; Bashammakh, S.; Hörtnagl, H.; Voits, M.; Fink, H.; Bader, M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 2003, 299, 76. [Google Scholar] [CrossRef]
- Mockus, S.M.; Vrana, K.E. Advances in the molecular characterization of tryptophan hydroxylase. J. Mol. Neurosci. 1998, 10, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Cornide-Petronio, M.E.; Anadón, R.; Rodicio, M.C.; Barreiroiglesias, A. The sea lamprey tryptophan hydroxylase: New insight into the evolution of the serotonergic system of vertebrates. Brain Struct. Funct. 2013, 218, 587–593. [Google Scholar] [CrossRef]
- Lillesaar, C. The serotonergic system in fish. J. Chem. Neuroanat. 2011, 41, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Peer, Y.V.D. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stangethomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozoufcostaz, C.; Bernot, A. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946. [Google Scholar] [CrossRef] [PubMed]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [PubMed]
- Bellipanni, G.; Rink, E.; Ballycuif, L. Cloning of two tryptophan hydroxylase genes expressed in the diencephalon of the developing zebrafish brain. Gene Expr. Patterns 2002, 119, S215–S220. [Google Scholar] [CrossRef]
- Varadarajan, N.; Gam, J.; Olsen, M.J.; Georgiou, G.; Iverson, B.L. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc. Natl. Acad. Sci. USA 2005, 102, 6855–6860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkman, T.J.; Martins, T.R.; Sutton, E.; Stout, J.T. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol. Biol. Evol. 2007, 24, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef] [Green Version]
- Smith, N.G.C.; Eyre-Walker, A. Adaptive protein evolution in Drosophila. Nature 2002, 415, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.G.; Dean, A.M.; Mitchellolds, T. Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef] [Green Version]
- Kajitani, R.; Toshimoto, K.; Noguchi, H.; Toyoda, A.; Ogura, Y.; Okuno, M.; Yabana, M.; Harada, M.; Nagayasu, E.; Maruyama, H. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.; Hill, C.M.; Wu, S.; Ruan, J.; Ma, Z.S. Dbg2olc: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 2016, 6, 31900. [Google Scholar] [CrossRef]
- Mount, D.W. Using the basic local alignment search tool (blast). CSH Protoc. 2007, 2007. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Prottest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using latex2e. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic. Acids. Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delano, W.L. Pymol: An open-source molecular graphics tool. CCP4 News. On Prot. Crys. 2002, 40, 82–92. [Google Scholar]
- Yang, Z. Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Carkaci-Salli, N.; Flanagan, J.M.; Martz, M.K.; Salli, U.; Walther, D.J.; Bader, M.; Vrana, K.E. Functional domains of human tryptophan hydroxylase 2 (htph2). J. Biol. Chem. 2006, 281, 28105–28112. [Google Scholar] [CrossRef] [PubMed]
- Winge, I.; McKinney, J.A.; Ying, M.; D’Santos, C.S.; Kleppe, R.; Knappskog, P.M.; Haavik, J. Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding. Biochem. J. 2008, 410, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Wang, X.R.; Zeng, Q.Y. Structural and functional evolution of positively selected sites in pine glutathione s-transferase enzyme family. J. Biol. Chem. 2013, 288, 24441–24451. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 2012, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045. [Google Scholar] [CrossRef]
- Meyer, A.; Van de Peer, Y. From 2r to 3r: Evidence for a fish-specific genome duplication (fsgd). Bioessays 2005, 27, 937–945. [Google Scholar] [CrossRef]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [Green Version]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Ludwig, A.; Wang, D.; Diogo, R.; Wei, Q.; He, S. Age and biogeography of major clades in sturgeons and paddlefishes (pisces: Acipenseriformes). Mol. Phylogenet. Evol. 2007, 42, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Khokha, M.K.; Grammer, T.C.; Harland, R.M.; Richardson, P.; Rokhsar, D.S. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, Y.; Bian, C.; You, X.; Deng, L.; Shi, Q. A comparative genomic survey provides novel insights into molecular evolution of l-aromatic amino acid decarboxylase in vertebrates. Molecules 2018, 23, 917. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, X.; Bian, C.; Yu, H.; Coon, S.L.; Shi, Q. Molecular evolution of aralkylamine N-acetyltransferase in fish: A genomic survey. Int. J. Mol. Sci. 2015, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ruan, Z.; Li, J.; Bian, C.; You, X.; Coon, S.L.; Shi, Q. A comparative genomic and transcriptomic survey provides novel insights into N-acetylserotonin methyltransferase (asmt) in fish. Molecules 2017, 22, 1653. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef]
- Ravi, V.; Bhatia, S.; Gautier, P.; Loosli, F.; Tay, B.H.; Tay, A.; Murdoch, E.; Coutinho, P.; van Heyningen, V.; Brenner, S.; et al. Sequencing of pax6 loci from the elephant shark reveals a family of pax6 genes in vertebrate genomes, forged by ancient duplications and divergences. PLoS Genet. 2013, 9, e1003177. [Google Scholar] [CrossRef]
- Yoshikuni, Y.; Ferrin, T.E.; Keasling, J.D. Designed divergent evolution of enzyme function. Nature 2006, 440, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, H.; Moran, Y.; Gordon, D.; Turkov, M.; Kahn, R.; Gurevitz, M. Positions under positive selection--key for selectivity and potency of scorpion α-toxins. Mol. Biol. Evol. 2009, 27, 1025. [Google Scholar] [CrossRef] [PubMed]
- Huber, R. Flexibility and rigidity of proteins and protein–pigment complexes. Angew. Chem. Int. Ed. 2010, 27, 79–88. [Google Scholar] [CrossRef]
- Zhang, J.; Dean, A.M.; Brunet, F.; Long, M. Evolving protein functional diversity in new genes of drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 16246–16250. [Google Scholar] [CrossRef] [PubMed]
- Nocito, A.; Dahm, F.; Jochum, W.; Jang, J.H.; Georgiev, P.; Bader, M.; Renner, E.L.; Clavien, P.A. Serotonin mediates oxidative stress and mitochondrial toxicity in a murine model of nonalcoholic steatohepatitis. Gastroenterology 2007, 133, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Marzioni, M.; Glaser, S.; Francis, H.; Marucci, L.; Benedetti, A.; Alvaro, D.; Taffetani, S.; Ueno, Y.; Roskams, T.; Phinizy, J.L. Autocrine/paracrine regulation of the growth of the biliary tree by the neuroendocrine hormone serotonin. Gastroenterology 2005, 128, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Lesurtel, M.; Graf, R.; Aleil, B.; Walther, D.J.; Tian, Y.; Jochum, W.; Gachet, C.; Bader, M.; Clavien, P.-A. Platelet-derived serotonin mediates liver regeneration. Science 2006, 312, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Nocito, A.; Georgiev, P.; Dahm, F.; Jochum, W.; Bader, M.; Graf, R.; Clavien, P.A. Platelets and platelet-derived serotonin promote tissue repair after normothermic hepatic ischemia in mice. Hepatology 2007, 45, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Tamim, H.; Shapiro, S.; Stang, M.R.; Collet, J.-P. Use of antidepressants and risk of colorectal cancer: A nested case-control study. Lancet Oncol. 2006, 7, 301–308. [Google Scholar] [CrossRef]
- Nocito, A.; Dahm, F.; Jochum, W.; Jang, J.H.; Georgiev, P.; Bader, M.; Graf, R.; Clavien, P.-A. Serotonin regulates macrophage-mediated angiogenesis in a mouse model of colon cancer allografts. Cancer Res. 2008, 68, 5152–5158. [Google Scholar] [CrossRef]
Sample Availability: Not available. |








| Class | Species Number | tph1 (tph1a) | tph1b | tph2 |
|---|---|---|---|---|
| Mammals | 11 | 11 | 0 | 11 |
| Birds | 18 | 18 | 0 | 18 |
| Reptiles | 9 | 9 | 0 | 9 |
| Amphibians | 3 | 4 | 0 | 4 |
| Actinopterygii | 29 | 37 | 26 | 37 |
| Total | 70 | 79 | 26 | 79 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Li, Y.; Lv, Y.; Bian, C.; You, X.; Endoh, D.; Teraoka, H.; Shi, Q. Molecular Evolution of Tryptophan Hydroxylases in Vertebrates: A Comparative Genomic Survey. Genes 2019, 10, 203. https://doi.org/10.3390/genes10030203
Xu J, Li Y, Lv Y, Bian C, You X, Endoh D, Teraoka H, Shi Q. Molecular Evolution of Tryptophan Hydroxylases in Vertebrates: A Comparative Genomic Survey. Genes. 2019; 10(3):203. https://doi.org/10.3390/genes10030203
Chicago/Turabian StyleXu, Junmin, Yanping Li, Yunyun Lv, Chao Bian, Xinxin You, Daiji Endoh, Hiroki Teraoka, and Qiong Shi. 2019. "Molecular Evolution of Tryptophan Hydroxylases in Vertebrates: A Comparative Genomic Survey" Genes 10, no. 3: 203. https://doi.org/10.3390/genes10030203





