Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Conserved Signature Indels and Phylogenetic Analysis
2.2. Homology Modelling and Structural Analyses
3. Results
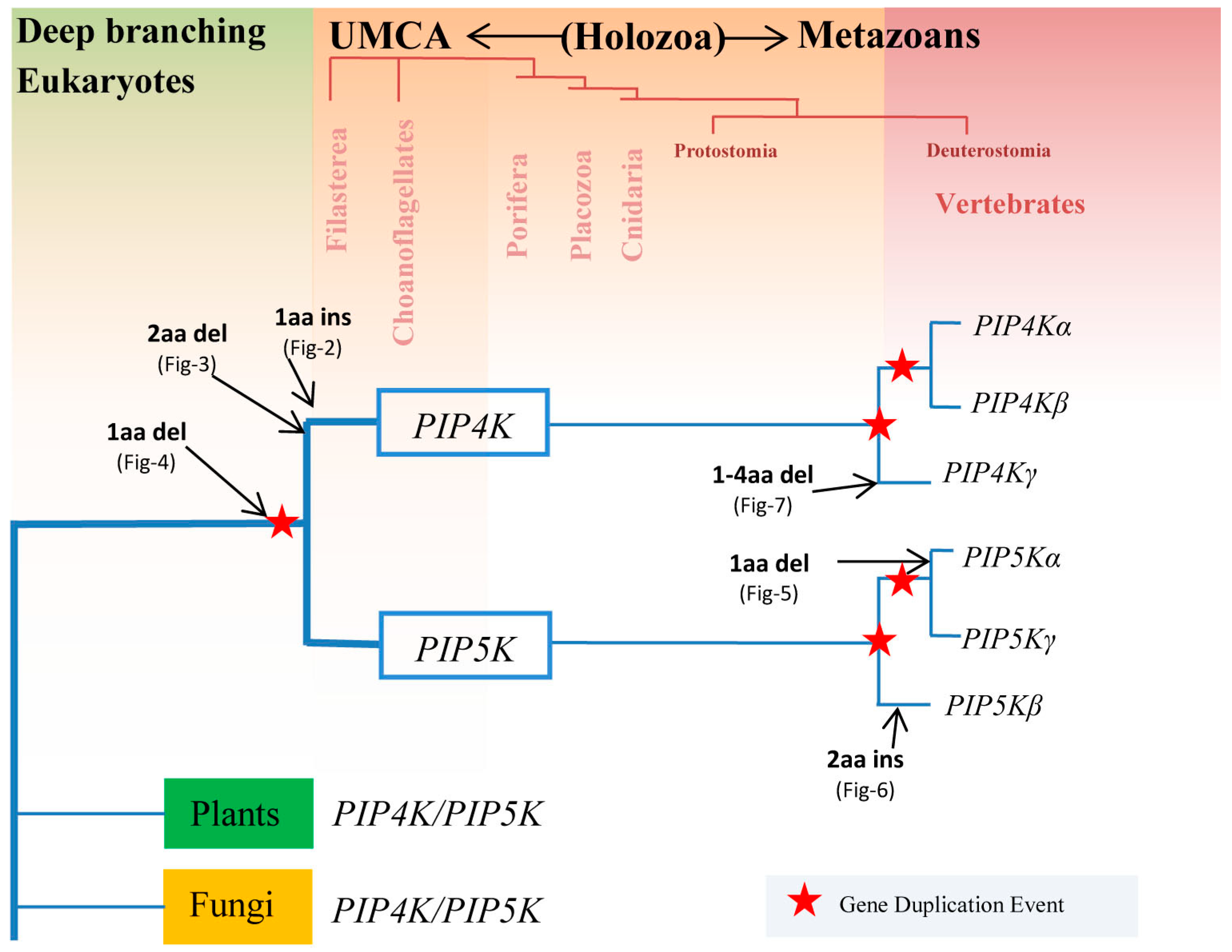
3.1. Species Distribution and Phylogenetic Analysis of PIP4K/PIP5K Protein Family
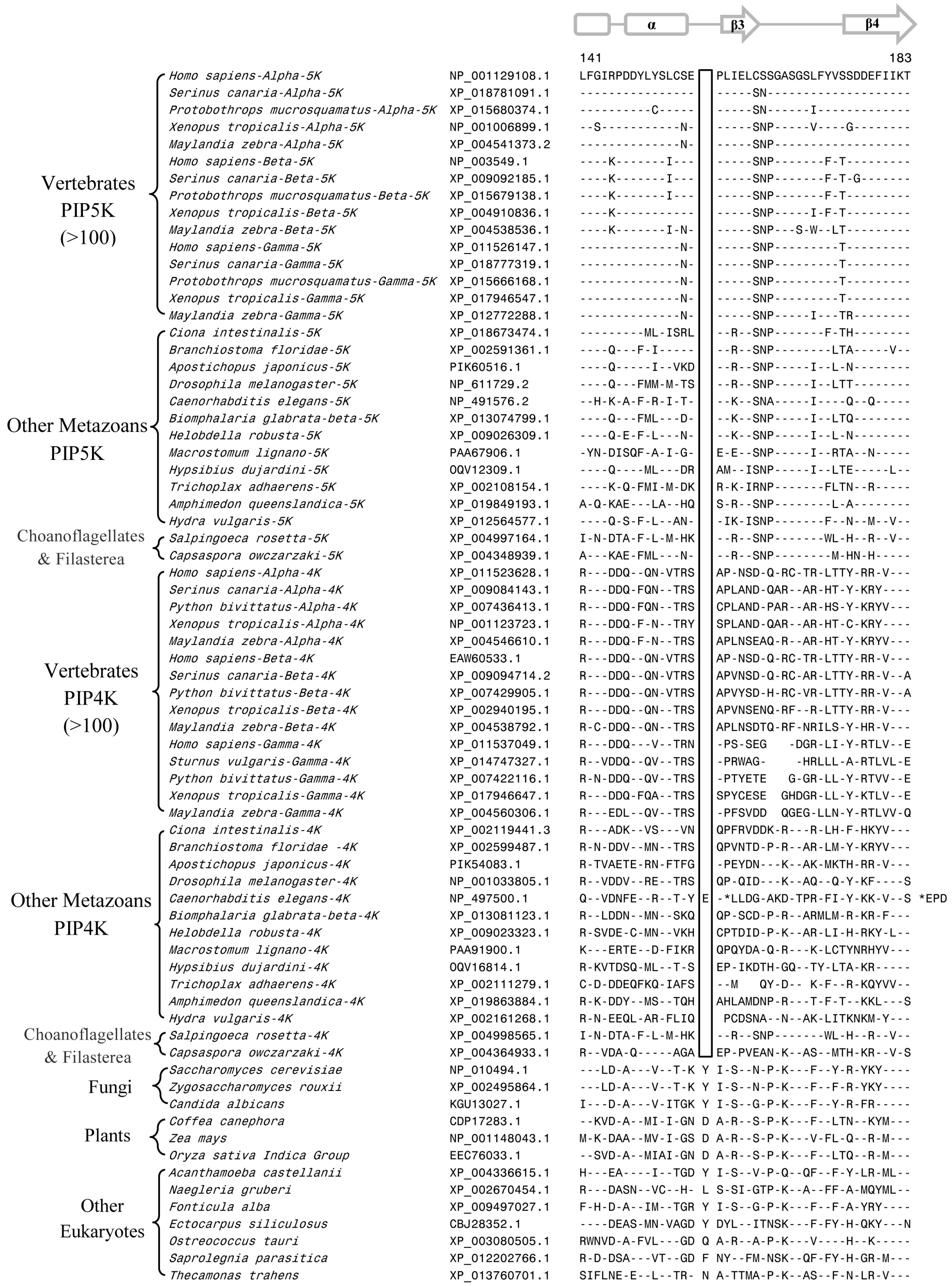
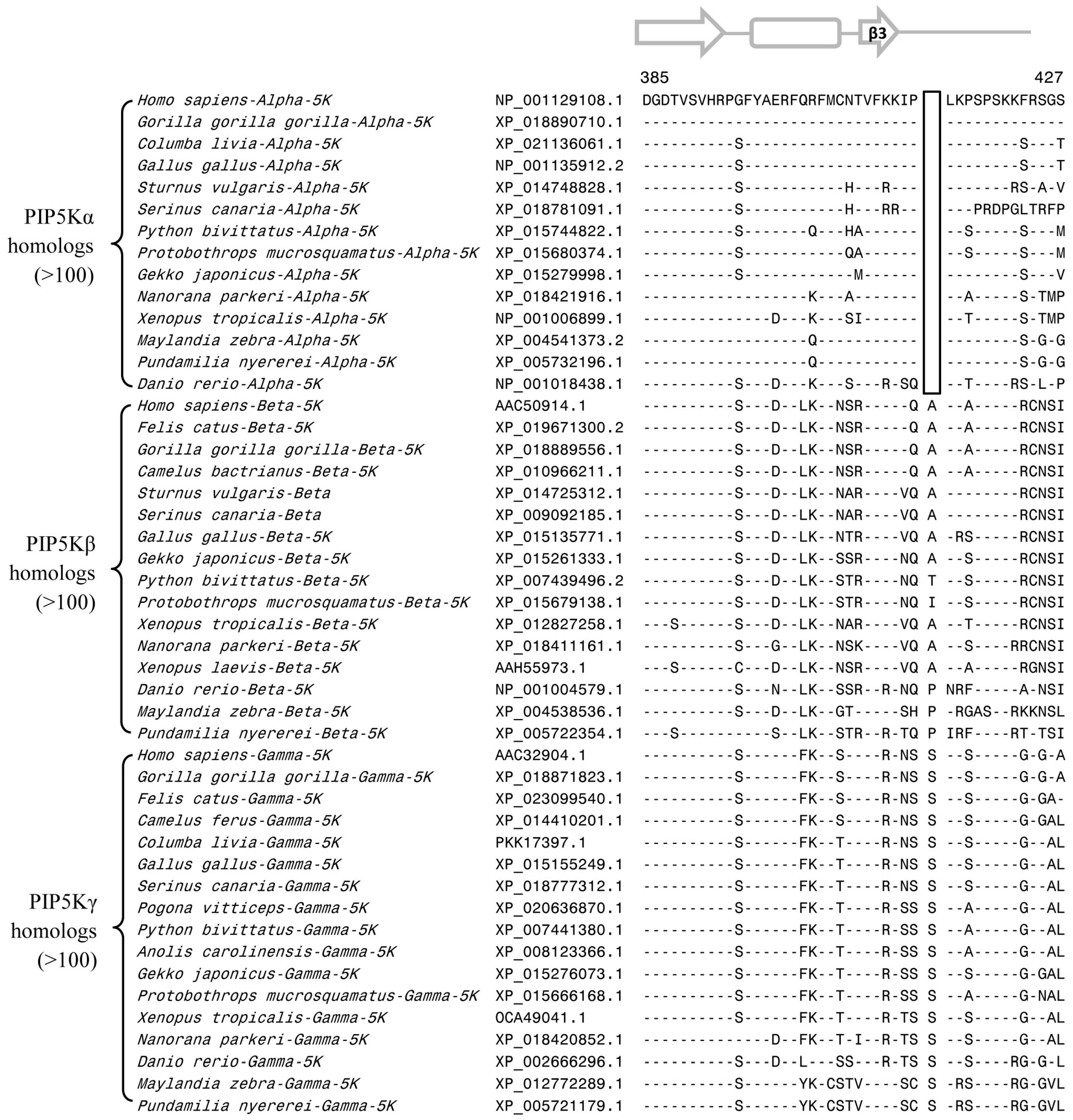
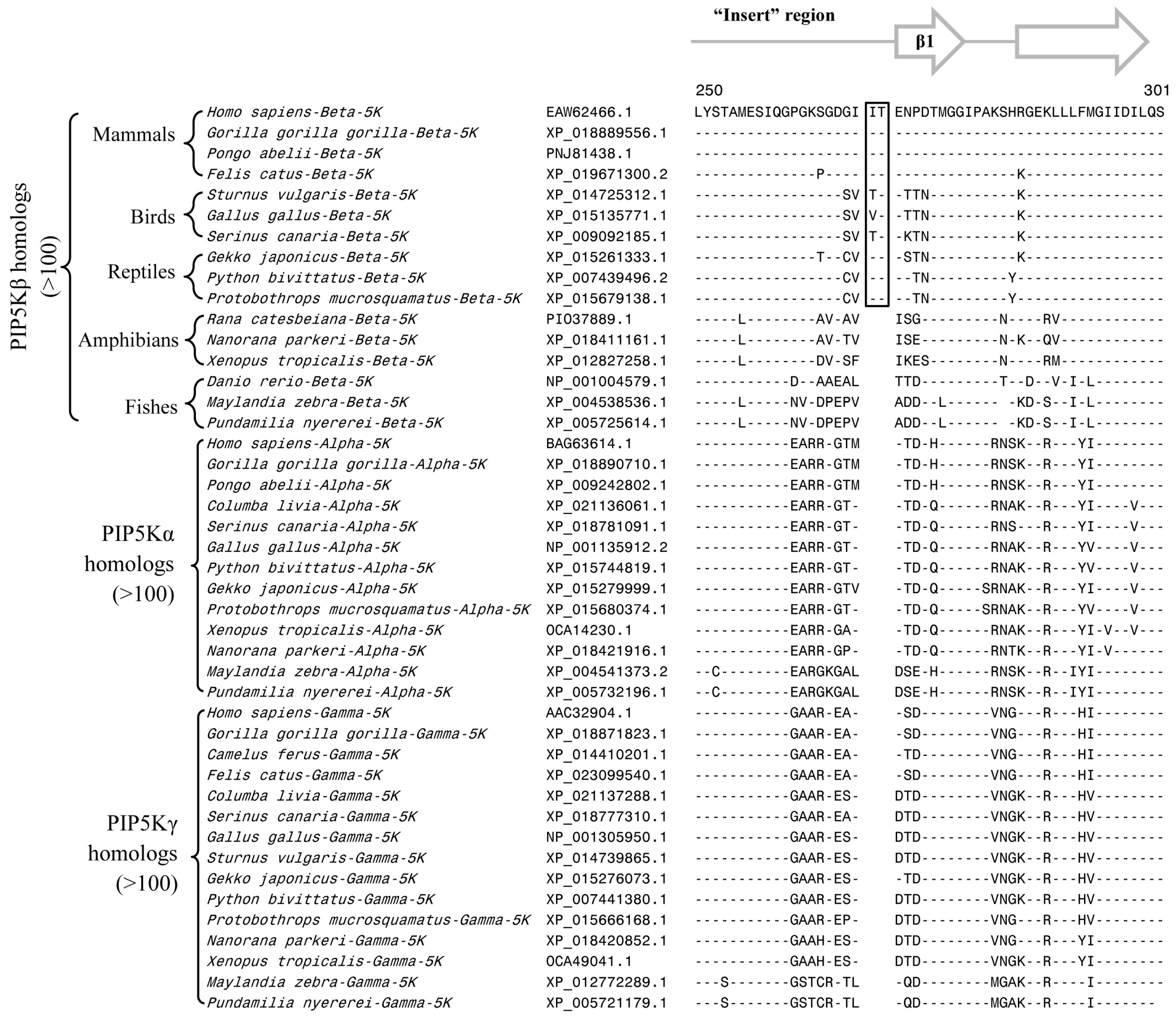
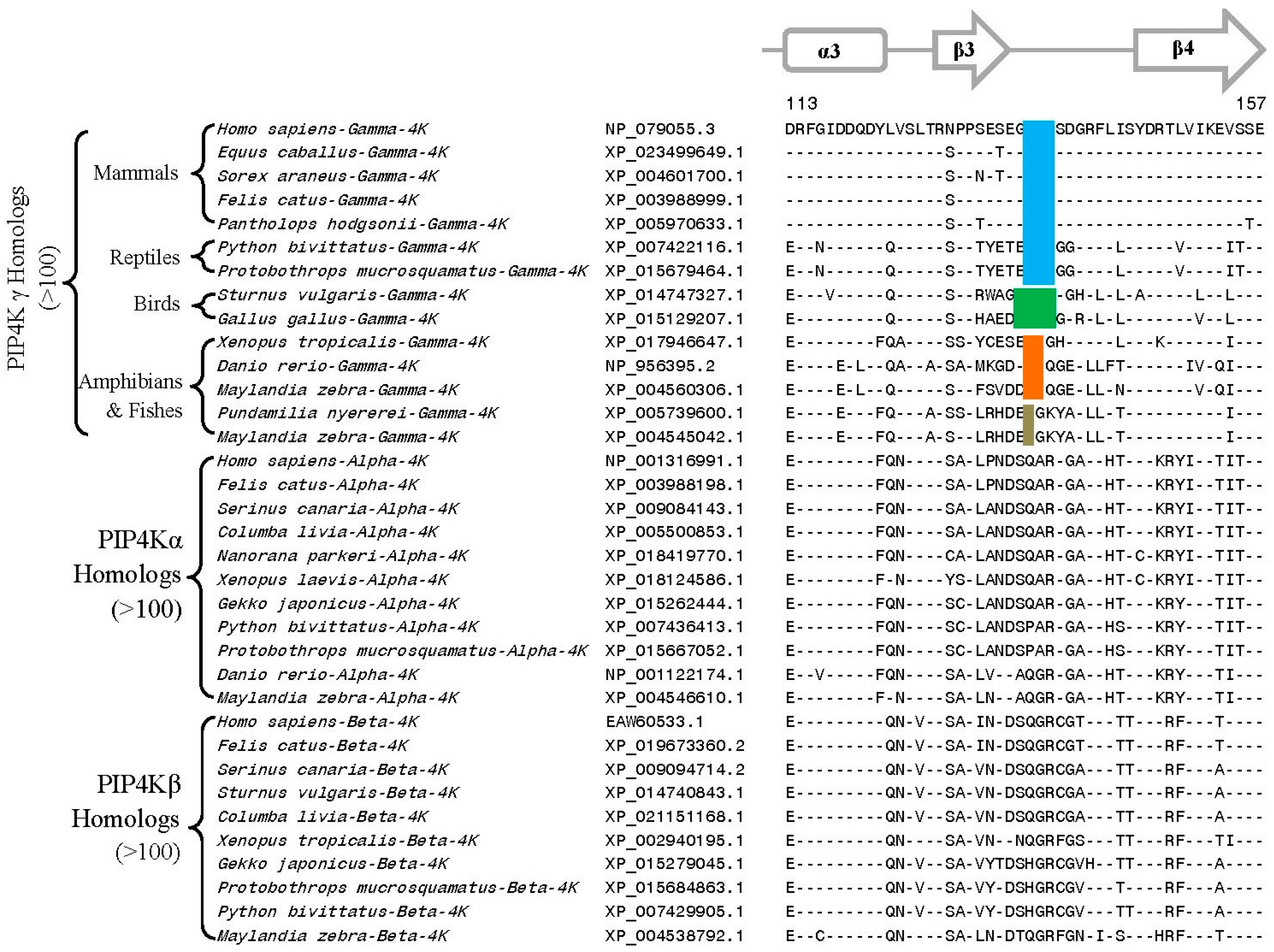
3.2. Conserved Signature Indels that are Distinctive Features of the PIP4K and PIP5K Family of Proteins and the Insights Provided by Them into the Evolutionary Relationships
3.3. Locations of the Identified CSIs in the Structures of the PIP4K/PIP5K Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef]
- Balla, T.; Szentpetery, Z.; Kim, Y.J. Phosphoinositide signaling: New tools and insights. Physiology 2009, 24, 231–244. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef]
- Brown, J.R.; Auger, K.R. Phylogenomics of phosphoinositide lipid kinases: Perspectives on the evolution of second messenger signaling and drug discovery. BMC. Evol. Biol. 2011, 11, 4. [Google Scholar] [CrossRef]
- Loijens, J.C.; Boronenkov, I.V.; Parker, G.J.; Anderson, R.A. The phosphatidylinositol 4-phosphate 5-kinase family. Adv. Enzyme Regul. 1996, 36, 115–140. [Google Scholar] [CrossRef]
- Aikawa, Y.; Martin, T.F. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J. Cell. Biol. 2003, 162, 647–659. [Google Scholar] [CrossRef] [Green Version]
- Mellman, D.L.; Gonzales, M.L.; Song, C.; Barlow, C.A.; Wang, P.; Kendziorski, C.; Anderson, R.A. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 2008, 451, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, H.; Shibasaki, Y.; Kizuki, N.; Wada, T.; Yazaki, Y.; Asano, T.; Oka, Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 1998, 273, 8741–8748. [Google Scholar]
- Ishihara, H.; Shibasaki, Y.; Kizuki, N.; Katagiri, H.; Yazaki, Y.; Asano, T.; Oka, Y. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J. Biol. Chem. 1996, 271, 23611–23614. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.H.; Emson, P.C.; Irvine, R.F. Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am. J. Physiol. Renal. Physiol. 2008, 295, F1422–F1430. [Google Scholar] [CrossRef]
- Clarke, J.H.; Irvine, R.F. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 2013, 454, 49–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Irvine, R.F.; Giudici, M.L. Phosphatidylinositol 4-phosphate 5-kinase Igamma_v6, a new splice variant found in rodents and humans. Biochem. Biophys. Res. Commun. 2011, 411, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Shulga, Y.V.; Anderson, R.A.; Topham, M.K.; Epand, R.M. Phosphatidylinositol-4-phosphate 5-kinase isoforms exhibit acyl chain selectivity for both substrate and lipid activator. J. Biol. Chem. 2012, 287, 35953–35963. [Google Scholar] [CrossRef] [PubMed]
- Desrivieres, S.; Cooke, F.T.; Parker, P.J.; Hall, M.N. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 15787–15793. [Google Scholar] [CrossRef] [PubMed]
- Guillas, I.; Vernay, A.; Vitagliano, J.J.; Arkowitz, R.A. Phosphatidylinositol 4,5-bisphosphate is required for invasive growth in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 3602–3614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okazaki, K.; Miyagishima, S.Y.; Wada, H. Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 2015, 27, 663–674. [Google Scholar] [CrossRef]
- Heilmann, I. Phosphoinositide signaling in plant development. Development 2016, 143, 2044–2055. [Google Scholar] [CrossRef] [Green Version]
- Mueller-Roeber, B.; Pical, C. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 2002, 130, 22–46. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef] [Green Version]
- Shisheva, A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol. Int. 2008, 32, 591–604. [Google Scholar] [CrossRef] [Green Version]
- Oude Weernink, P.A.; Schmidt, M.; Jakobs, K.H. Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 2004, 500, 87–99. [Google Scholar] [CrossRef]
- Van den Bout, I.; Divecha, N. PIP5K-driven PtdIns(4,5)P2 synthesis: Regulation and cellular functions. J. Cell Sci. 2009, 122, 3837–3850. [Google Scholar] [CrossRef]
- Bulley, S.J.; Clarke, J.H.; Droubi, A.; Giudici, M.L.; Irvine, R.F. Exploring phosphatidylinositol 5-phosphate 4-kinase function. Adv. Biol. Regul. 2015, 57, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Baldauf, S.L.; Palmer, J.D. Animals and fungi are each other’s closest relatives: Congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 11558–11562. [Google Scholar] [CrossRef]
- Gupta, R.S.; Aitken, K.; Falah, M.; Singh, B. Cloning of Giardia lamblia heat shock protein HSP70 homologs: Implications regarding origin of eukaryotic cells and of endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1994, 91, 2895–2899. [Google Scholar] [CrossRef]
- Gupta, R.S. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 1998, 62, 1435–1491. [Google Scholar] [PubMed]
- Gupta, R.S. Molecular signatures that are distinctive characteristics of the vertebrates and chordates and supporting a grouping of vertebrates with the tunicates. Mol. Phylogenet. Evol. 2016, 94, 383–391. [Google Scholar] [CrossRef]
- Rivera, M.C.; Lake, J.A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 1992, 257, 74–76. [Google Scholar] [CrossRef]
- Rokas, A.; Holland, P.W. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000, 15, 454–459. [Google Scholar] [CrossRef]
- Gupta, R.S.; Epand, R.M. Phylogenetic analysis of the diacylglycerol kinase family of proteins and identification of multiple highly-specific conserved inserts and deletions within the catalytic domain that are distinctive characteristics of different classes of DGK homologs. PLoS ONE 2017, 12, e0182758. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017, 45, D12–D17. [Google Scholar] [CrossRef]
- Methods in Microbiology New Approaches to Prokaryotics Systematics; Goodfellow, I.C.S.M.; Chun, J. (Eds.) Academic Press: London, UK, 2014; pp. 153–182. [Google Scholar]
- Khadka, B.; Gupta, R.S. Identification of a conserved 8 aa insert in the PIP5K protein in the Saccharomycetaceae family of fungi and the molecular dynamics simulations and structural analysis to investigate its potential functional role. Proteins 2017, 85, 1454–1467. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.S. Impact of genomics on the understanding of microbial evolution and classification: The importance of Darwin’s views on classification. FEMS Microbiol. Rev. 2016, 40, 520–553. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Gupta, R.S.; Nanda, A.; Khadka, B. Novel molecular, structural and evolutionary characteristics of the phosphoketolases from bifidobacteria and Coriobacteriales. PLoS ONE 2017, 12, e0172176. [Google Scholar] [CrossRef]
- Khadka, B.; Adeolu, M.; Blankenship, R.E.; Gupta, R.S. Novel insights into the origin and diversification of photosynthesis based on analyses of conserved indels in the core reaction center proteins. Photosynth. Res. 2017, 131, 159–171. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlic, A.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Westbrook, J.D.; Woo, J.; et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015, 43, D345–D356. [Google Scholar] [CrossRef]
- Hu, J.; Yuan, Q.; Kang, X.; Qin, Y.; Li, L.; Ha, Y.; Wu, D. Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled. Nat. Commun. 2015, 6, 8205. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.D.; Misra, S.; Boronenkov, I.V.; Anderson, R.A.; Hurley, J.H. Structure of type IIbeta phosphatidylinositol phosphate kinase: A protein kinase fold flattened for interfacial phosphorylation. Cell 1998, 94, 829–839. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [Green Version]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007, 50, 1–4. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Benkert, P.; Tosatto, S.C.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins 2008, 71, 261–277. [Google Scholar] [CrossRef]
- Mendoza, L.; Taylor, J.W.; Ajello, L. The class mesomycetozoea: A heterogeneous group of microorganisms at the animal-fungal boundary. Annu. Rev. Microbiol. 2002, 56, 315–344. [Google Scholar] [CrossRef] [PubMed]
- Shalchian-Tabrizi, K.; Minge, M.A.; Espelund, M.; Orr, R.; Ruden, T.; Jakobsen, K.S.; Cavalier-Smith, T. Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE 2008, 3, e2098. [Google Scholar] [CrossRef]
- Torruella, G.; Derelle, R.; Paps, J.; Lang, B.F.; Roger, A.J.; Shalchian-Tabrizi, K.; Ruiz-Trillo, I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol. Biol. Evol. 2012, 29, 531–544. [Google Scholar] [CrossRef]
- Hehenberger, E.; Tikhonenkov, D.V.; Kolisko, M.; del Campo, J.; Esaulov, A.S.; Mylnikov, A.P.; Keeling, P.J. Novel Predators Reshape Holozoan Phylogeny and Reveal the Presence of a Two-Component Signaling System in the Ancestor of Animals. Curr. Biol. 2017, 27, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Stanhope, M.J.; Madsen, O.; de Jong, W.W. Molecules consolidate the placental mammal tree. Trends Ecol. Evol. 2004, 19, 430–438. [Google Scholar] [CrossRef]
- Carr, M.; Leadbeater, B.S.; Hassan, R.; Nelson, M.; Baldauf, S.L. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc. Natl. Acad. Sci. USA 2008, 105, 16641–16646. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Trillo, I.; Roger, A.J.; Burger, G.; Gray, M.W.; Lang, B.F. A phylogenomic investigation into the origin of metazoa. Mol. Biol. Evol. 2008, 25, 664–672. [Google Scholar] [CrossRef]
- Steenkamp, E.T.; Wright, J.; Baldauf, S.L. The protistan origins of animals and fungi. Mol. Biol. Evol. 2006, 3, 93–106. [Google Scholar] [CrossRef]
- Lang, B.F.; O’Kelly, C.; Nerad, T.; Gray, M.W.; Burger, G. The closest unicellular relatives of animals. Curr. Biol. 2002, 12, 1773–1778. [Google Scholar] [CrossRef]
- Kumar, S.; Hedges, S.B. A molecular timescale for vertebrate evolution. Nature 1998, 392, 917–920. [Google Scholar] [CrossRef]
- Akiva, E.; Itzhaki, Z.; Margalit, H. Built-in loops allow versatility in domain-domain interactions: Lessons from self-interacting domains. Proc. Natl. Acad. Sci. USA 2008, 105, 13292–13297. [Google Scholar] [CrossRef]
- Hashimoto, K.; Panchenko, A.R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. USA 2010, 107, 20352–20357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alnajar, S.; Khadka, B.; Gupta, R.S. Ribonucleotide Reductases from Bifidobacteria Contain Multiple Conserved Indels Distinguishing Them from All Other Organisms: In Silico Analysis of the Possible Role of a 43 aa Bifidobacteria-Specific Insert in the Class III RNR Homolog. Front. Microbiol. 2017, 8, 1409. [Google Scholar] [CrossRef]
- Hassan, F.M.N.; Gupta, R.S. Novel Sequence Features of DNA Repair Genes/Proteins from Deinococcus Species Implicated in Protection from Oxidatively Generated Damage. Genes 2018, 9, 149. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.L.; Prajda, N.; Yeh, Y.A.; Weber, G. 1-Phosphatidylinositol 4-phosphate 5-kinase (EC 2.7.1.68): A proliferation- and malignancy-linked signal transduction enzyme. Cancer Res. 1994, 54, 5574–5578. [Google Scholar]
- Luoh, S.W.; Venkatesan, N.; Tripathi, R. Overexpression of the amplified Pip4k2beta gene from 17q11–12 in breast cancer cells confers proliferation advantage. Oncogene 2004, 23, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Bourdeaut, F.; Combaret, V.; Picrron, G.; Raynal, V.; Aurias, A.; Ribeiro, A.; Janoueix-Lerosey, I.; Delattre, O. Stepwise occurrence of a complex unbalanced translocation in neuroblastoma leading to insertion of a telomere sequence and late chromosome 17q gain. Oncogene 2005, 24, 3377–3384. [Google Scholar] [CrossRef] [Green Version]
- Narkis, G.; Ofir, R.; Landau, D.; Manor, E.; Volokita, M.; Hershkowitz, R.; Elbedour, K.; Birk, O.S. Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI gamma of the phophatidylinsitol pathway. Am. J. Hum. Genet. 2007, 81, 530–539. [Google Scholar] [CrossRef]
- Wang, Y.; Lian, L.; Golden, J.A.; Morrisey, E.E.; Abrams, C.S. PIP5KI gamma is required for cardiovascular and neuronal development. Proc. Natl. Acad. Sci. USA 2007, 104, 11748–11753. [Google Scholar] [CrossRef]
- Porciello, N.; Kunkl, M.; Viola, A.; Tuosto, L. Phosphatidylinositol 4-Phosphate 5-Kinases in the Regulation of T Cell Activation. Front. Immunol. 2016, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Emerling, B.M.; Hurov, J.B.; Poulogiannis, G.; Tsukazawa, K.S.; Choo-Wing, R.; Wulf, G.M.; Bell, E.L.; Shim, H.S.; Lamia, K.A.; Rameh, L.E.; et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 2013, 155, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Semenas, J.; Hedblom, A.; Miftakhova, R.R.; Sarwar, M.; Larsson, R.; Shcherbina, L.; Johansson, M.E.; Harkonen, P.; Sterner, O.; Persson, J.L. The role of PI3K/AKT-related PIP5K1alpha and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E3689–E3698. [Google Scholar] [CrossRef] [PubMed]
- Wright, B.D.; Simpson, C.; Stashko, M.; Kireev, D.; Hull-Ryde, E.A.; Zylka, M.J.; Janzen, W.P. Development of a High-Throughput Screening Assay to Identify Inhibitors of the Lipid Kinase PIP5K1C. J. Biomol. Screen. 2015, 20, 655–662. [Google Scholar] [CrossRef]
- Voss, M.D.; Czechtizky, W.; Li, Z.; Rudolph, C.; Petry, S.; Brummerhop, H.; Langer, T.; Schiffer, A.; Schaefer, H.L. Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem. Biophys. Res. Commun. 2014, 449, 327–331. [Google Scholar] [CrossRef]
- Hayakawa, N.; Noguchi, M.; Takeshita, S.; Eviryanti, A.; Seki, Y.; Nishio, H.; Yokoyama, R.; Noguchi, M.; Shuto, M.; Shima, Y.; et al. Structure-activity relationship study, target identification, and pharmacological characterization of a small molecular IL-12/23 inhibitor, APY0201. Bioorg. Med. Chem. 2014, 22, 3021–3029. [Google Scholar] [CrossRef]
- Philippon, H.; Brochier-Armanet, C.; Perriere, G. Evolutionary history of phosphatidylinositol- 3-kinases: Ancestral origin in eukaryotes and complex duplication patterns. BMC. Evol. Biol. 2015, 15, 226. [Google Scholar] [CrossRef]
- Singh, B.; Gupta, R.S. Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Genet. Genom. 2009, 281, 361–373. [Google Scholar] [CrossRef]
- Gao, B.; Gupta, R.S. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 66–112. [Google Scholar] [CrossRef]




| Taxa/Phylum | PIP4K | PIP5K | PIP4K/PIP5K | ||
|---|---|---|---|---|---|
| DEUTEROSTOMIA | Vertebrates | Mammals | >100 | >100 | - |
| Birds | >100 | >100 | - | ||
| Amphibians | 4 | 4 | - | ||
| Reptiles | 10 | 10 | - | ||
| Fishes | >50 | >50 | - | ||
| Tunicata | 2 | 2 | - | ||
| Cephalochordata | 2 | 2 | - | ||
| Hemichordata | 1 | 1 | - | ||
| Echinodermata | 1 | 1 | - | ||
| PROTOSTOMIA | Arthropoda | >100 | >100 | - | |
| Nematoda | >50 | >50 | - | ||
| Mollusca | 4 | 4 | - | ||
| Annelida | 2 | 2 | - | ||
| Platyhelminthes | 2 | 2 | - | ||
| Tardigrada | 2 | 2 | - | ||
| Early Metazoans | PLACOZOA | 1 | 1 | - | |
| PORIFERA | 1 | 1 | - | ||
| CNIDARIA | 3 | 3 | - | ||
| UMCA | CHOANOFLAGELLATEA | 2 | 2 | - | |
| FILASTEREA | 1 | 1 | - | ||
| ICHTHYOSPOREA | 1 * | 1 * | - | ||
| FUNGI | - | - | >100 | ||
| PLANTS | - | - | >100 | ||
| OTHERS EUKARYOTES | APICOMPLEXA | - | - | >10 | |
| AMOEBOZOA | - | - | |||
| PERCOLOZOA (Naegleria gruberi) | - | - | |||
| APUSOZOA (Thecamonas trahens) | - | - | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadka, B.; Gupta, R.S. Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family. Genes 2019, 10, 312. https://doi.org/10.3390/genes10040312
Khadka B, Gupta RS. Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family. Genes. 2019; 10(4):312. https://doi.org/10.3390/genes10040312
Chicago/Turabian StyleKhadka, Bijendra, and Radhey S. Gupta. 2019. "Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family" Genes 10, no. 4: 312. https://doi.org/10.3390/genes10040312
APA StyleKhadka, B., & Gupta, R. S. (2019). Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family. Genes, 10(4), 312. https://doi.org/10.3390/genes10040312





