Genome-Wide Analysis of the miRNA–mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra
Abstract
1. Background
2. Methods
2.1. Plant Materials and Treatment
2.2. Total RNA Extraction and RNA Library Construction for Sequencing
2.3. Identification and Differential Expression Analysis of miRNAs
2.4. Analysis of Differential Expression Genes Based on Transcriptome Sequencing
2.5. GO and KEGG Pathway Enrichment Analysis
2.6. Quantitative Real-Time PCR Analysis
3. Results
3.1. Identification of miRNAs in Populus simonii × P. nigra
3.2. Differential Analysis of Candidate miRNAs Involved In Cold Tolerance in Populus simonii × P. nigra
3.3. Identification of Candidate Cold-Responsive Genes by Transcriptome Analysis
3.4. Regulation Analysis of miRNAs via Prediction of Target mRNA
3.5. Expression Analysis of miRNAs by qRT-PCR
3.6. qRT-PCR Validation of the Transcription Level of Target Genes
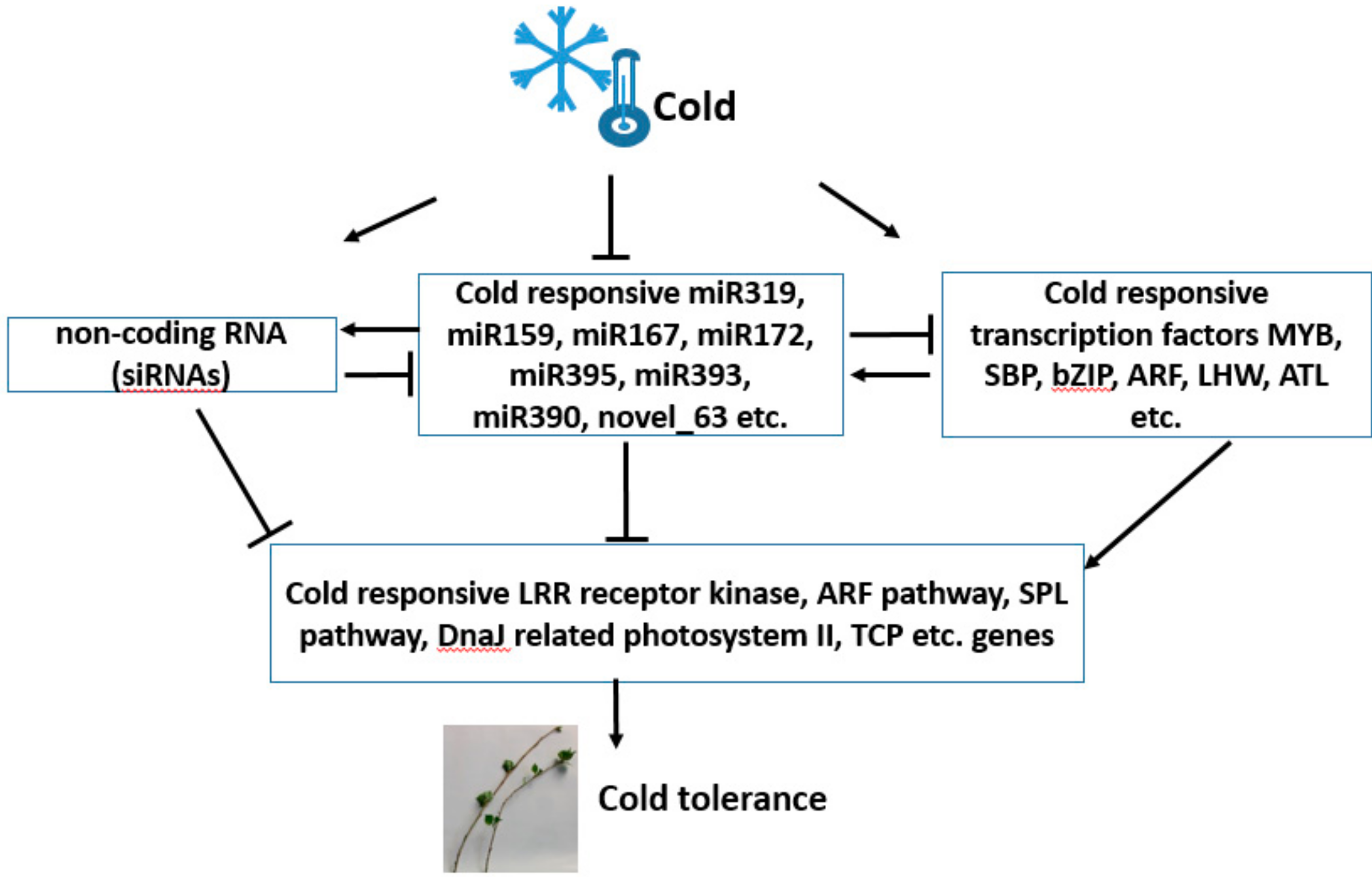
4. Discussion
4.1. Cold-Responsive miRNAs in Populus simonii × P. nigra
4.2. Differentially Expressed Transcripts under Cold Treatment
4.3. miRNAs May Be Involved in Cold Tolerance by Negatively Regulating Target mRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhao, J.G.; He, Q.S.; Chen, G.; Wang, L.; Jin, B. Regulation of Non-coding RNAs in Heat Stress Responses of Plants. Front. Plant Sci. 2016, 7, 1213. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.; Ren, Y.; Xu, J.; Zhang, Z.; Wang, Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 2012, 417, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.L. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, M.; Miura, S.; Nei, M. Origins and evolution of microRNA genes in plant species. Genome Biol. Evol. 2012, 4, 230–239. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Liu, R.; Lai, B.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Identification of microRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 7, 2059. [Google Scholar] [CrossRef]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, G.; Sutoh, K.; Zhu, J.K.; Zhang, W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 2008, 1779, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-responsive microRNAs in Populus. Plant J. 2008, 55, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.K.; Bai, X.; Li, Y.; Ding, X.D.; Ge, Y.; Cai, H.; Ji, W.; Wu, N.; Zhu, Y.M. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 2010, 459, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, X.; Chen, X.; Song, C.; Zou, Z.; Wang, Y.; Wang, M.; Fang, W.; Li, X. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014, 14, 271. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, F.; Zhang, Y.; Wang, L.; Cheng, Y. Cold-responsive miRNAs and their target genes in the wild eggplant species Solanum aculeatissimum. BMC Genom. 2017, 18, 1000. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, L.; Xu, C.; Yuan, S.; Zhang, F.; Zheng, Y.; Zhao, C. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012, 159, 721–738. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Song, G.; Zhang, R.; Zhang, S.; Li, Y.; Gao, J.; Han, X.; Chen, M.; Wang, J.; Li, W.; Li, G. Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genom. 2017, 18, 212. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Pan, Y.; Li, J.; Chen, X.; Pan, Y.; Wang, Y.; Tian, S.; Zhang, X. Heterologous expression of Arabidopsis C-repeat binding factor 3 (AtCBF3) and cold-regulated 15A (AtCOR15A) enhanced chilling tolerance in transgenic eggplant (Solanum melongena L.). Plant Cell Rep. 2014, 33, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.M.; Green, P.J.; et al. Criteria for Annotation of Plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef] [PubMed]
- Michael, Z. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar]
- Wu, H.J.; Ma, Y.K.; Chen, T.; Wang, M.; Wang, X.J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, M.B.; Tu, J.X.; Helliwell, C.A.; Waterhouse, P.M.; Dennis, E.S.; Fu, T.D.; Fan, Y.L. Cloning and characterization of microRNAs from Brassica napus. FEBS Lett. 2007, 581, 3848–3856. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Yang, B.; Zhang, A.; Ma, J.; Ding, Y.; Chen, Z.; Li, J.; Xu, X.; Liu, L. Genome-Wide Identification of MicroRNAs in Response to Cadmium Stress in Oilseed Rape (Brassica napus L.) Using High-Throughput Sequencing. Int. J. Mol. Sci. 2018, 19, 1431. [Google Scholar] [CrossRef]
- Barakat, A.; Sriram, A.; Park, J.; Zhebentyayeva, T.; Main, D.; Abbott, A. Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genom. 2012, 13, 481. [Google Scholar] [CrossRef]
- Niu, J.; Wang, J.; Hu, H.W.; Chen, Y.L.; An, J.Y.; Cai, J.; Sun, R.Z.; Sheng, Z.T.; Liu, X.P.; Lin, S.Z. Cross-talk between freezing response and signaling for regulatory transcriptions of MIR475b and its targets by miR475b promoter in Populus suaveolens. Sci. Rep. 2016, 6, 20648. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Rock, C.D. Trans-acting small interfering RNA4: Key to nutraceutical synthesis in grape development? Trends Plant Sci. 2013, 18, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, K.; Gao, P.; Liu, X.; Li, G.; Wu, Z. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 2014, 215, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, F.; Shiran, B.; Fallahi, H.; Mirakhorli, N.; Budak, H.; Martinez-Gomez, P. In silico search and biological validation of microRNAs related to drought response in peach and almond. Funct. Integr. Genom. 2017, 17, 189–201. [Google Scholar] [CrossRef]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis Transcription Factors SPL1 and SPL12 Confer Plant Thermotolerance at Reproductive Stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Kong, F.; Deng, Y.; Zhou, B.; Wang, G.; Wang, Y.; Meng, Q. A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 2014, 65, 143–158. [Google Scholar] [CrossRef]
- Thiebaut, F.; Rojas, C.A.; Almeida, K.L.; Grativol, C.; Domiciano, G.C.; Lamb, C.R.; Engler Jde, A.; Hemerly, A.S.; Ferreira, P.C. Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ. 2012, 35, 502–512. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Upadhyaya, N.M.; Gubler, F.; Helliwell, C.A. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009, 9, 149. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.M.; Li, X.W.; Zhou, Y.H.; Wang, F.W.; Wang, N.; Dong, Y.Y.; Yuan, Y.X.; Chen, H.; Liu, X.M.; Yao, N.; et al. The AaDREB1 Transcription Factor from the Cold-Tolerant Plant Adonis amurensis Enhances Abiotic Stress Tolerance in Transgenic Plant. Int. J. Mol. Sci. 2016, 17, 611. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Lang, Z.; Zhu, J.K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; An, C.S.; Hong, Y.N.; Lee, K.W. Cold-inducible transcription factor, CaCBF, is associated with a homeodomain leucine zipper protein in hot pepper (Capsicum annuum L.). Mol. Cells 2004, 18, 300–308. [Google Scholar] [PubMed]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Vo, K.; Danh Nguyen, C.; Jeong, D.-H.; Lee, S.-K.; Kumar, M.; Kim, S.-R.; Park, S.-H.; Kim, J.J.; Jeon, J.-S. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Zhuo, C.; Liang, L.; Zhao, Y.; Guo, Z.; Lu, S. A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 2018, 41, 2021–2032. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Chung, M.Y.; Hur, Y.; Cho, Y.G.; Watanabe, M.; Nou, I.S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhang, Z.G.; He, X.J.; Zhou, H.L.; Wen, Y.X.; Dai, J.X.; Zhang, J.S.; Chen, S.Y. A rice transcription factor OsbHLH1 is involved in cold stress response. Theor. Appl. Genet. 2003, 107, 1402–1409. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. 2017, 119, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.T.; Yu, Q.; Yang, Y.Y.; Su, Y.C.; Ahmad, W.; Guo, J.L.; Gao, S.W.; Xu, L.P.; Que, Y.X. Identification of cold-related miRNAs in sugarcane by small RNA sequencing and functional analysis of a cold inducible ScmiR393 to cold stress. Environ. Exp. Bot. 2018, 155, 464–476. [Google Scholar] [CrossRef]
- Zhang, S.L.; Wang, Y.N.; Li, K.X.; Zou, Y.M.; Chen, L.; Li, X. Identification of Cold-Responsive miRNAs and Their Target Genes in Nitrogen-Fixing Nodules of Soybean. Int. J. Mol. Sci. 2014, 15, 13596–13614. [Google Scholar] [CrossRef] [PubMed]
- Valiollahi, E.; Farsi, M.; Kakhki, A.M. Sly-miR166 and Sly-miR319 are components of the cold stress response in Solanum lycopersicum. Plant Biotechnol. Rep. 2014, 8, 349–356. [Google Scholar] [CrossRef]


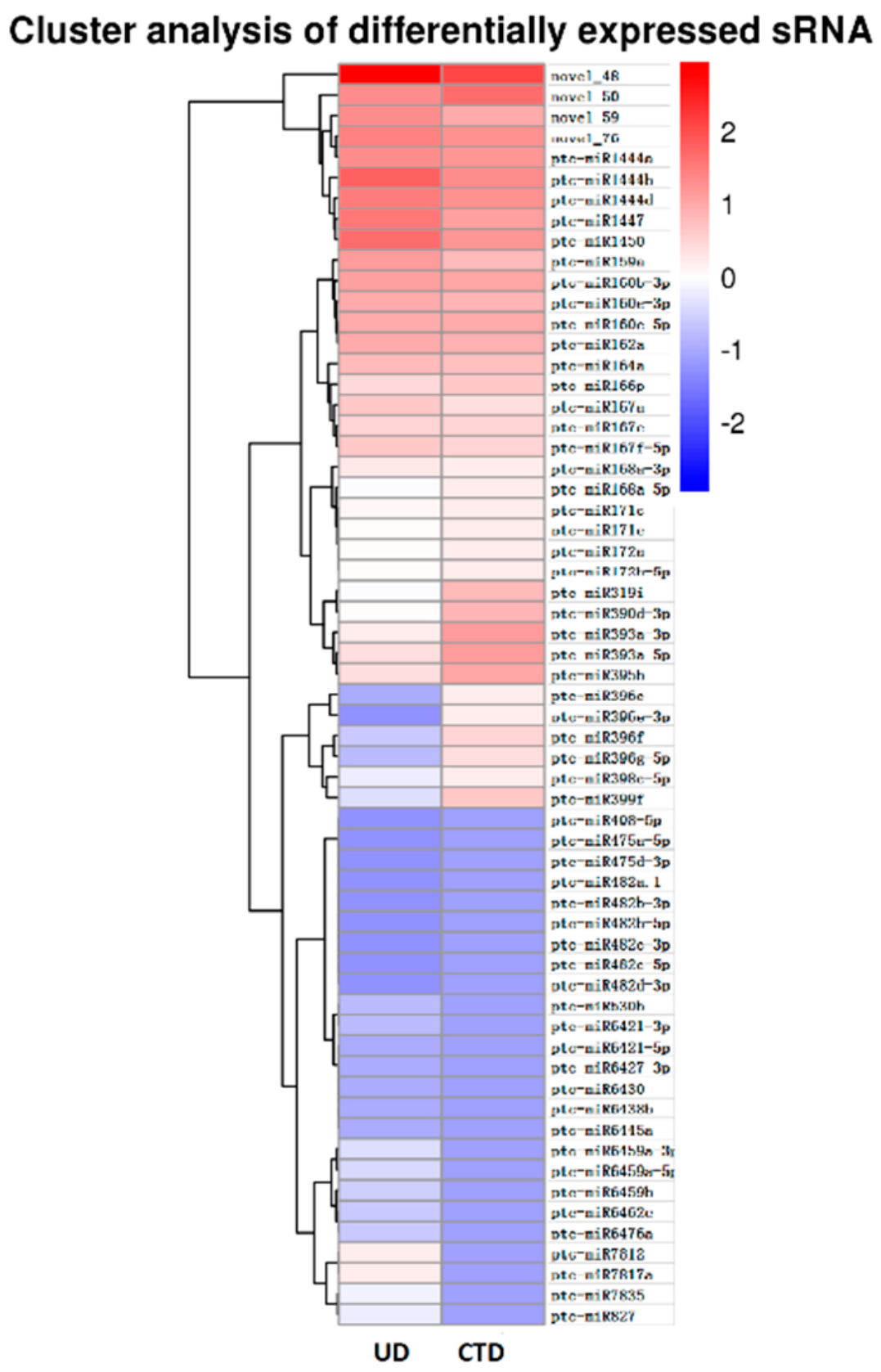




| Sample | Total Reads | Clean Reads | Total sRNA | Mapped sRNA | rRNA | snRNA | snoRNA | Ta-siRNA | Known miRNA | Novel miRNA |
|---|---|---|---|---|---|---|---|---|---|---|
| Cold Treated | 6,757,221 | 6,125,123 | 2,247,238 | 1,875,409 | 251,512 | 2012 | 19,535 | 235 | 10,089 | 118 |
| Untreated | 6,547,394 | 6,356,896 | 3,807,159 | 3,126,566 | 427,344 | 2570 | 34,948 | 2778 | 45,241 | 681 |
| Sample | Raw Reads | Clean Reads | Q20 (%) | Q30 (%) | Number of Transcripts (>200 bp) | N50 | N90 | Number of Genes (>200 bp) | N50 | N90 |
|---|---|---|---|---|---|---|---|---|---|---|
| CTD | 49,473,544 | 48,912,024 | 97.21 | 95.5 | 178,416 | 1347 | 266 | 92,755 | 1668 | 528 |
| UD | 55,000,000 | 54,357,974 | 97.17 | 95.46 |
| Target ID | Annotation | miRNA ID | Predicted Binding Site | Negative Regulation |
|---|---|---|---|---|
| Cluster-16493.31604 | PREDICTED: coatomer subunit alpha-1-like isoform X1 | novel_48 | miRNA: 1 TGTGGGAATGAACATTATGAG 21 | : | || :|||||| | | : : | : || : Target: s312 ATACCTTTACTTGTGGTGCTT 292 | yes |
| Cluster-16493.38601 | Putative EG45-like domain containing protein 1 | ptc-miR167e | miRNA: 1 TGAAGCTGCCAGCATGAT-CTG 21 |||| ||| : ||||| | *|| | **|| Target: 259 ACTTCGATGGTCGTCCTACAAC 238 | no |
| Cluster-16493.31604 | PREDICTED: coatomer subunit alpha-1-like isoform X1 | ptc-miR168a-5p | miRNA: 1 TCGCTTGGTGCAGGTCGGGAA 21 |* ||||| | | : ||: : ||| | | || Target: 3706 ACCGAACCATGTTTAGCCCTT 3686 | no |
| Cluster-16493.38601 | Putative EG45-like domain containing protein 1 | ptc-miR167f-5p | miRNA: 1 TGAAGCTGCCAGCATGATCTT 21 |||||| | : |||| | | *||| *|| Target: 259 ACTTCGATGGTCGTCCTACAA 239 | no |
| Cluster-16493.38601 | Putative EG45-like domain containing protein 1 | ptc-miR167a | miRNA: 1 TGAAGCTGCCAGCATGATCTA 21 ||||| || : ||| ||| * |||* |* Target: 259 ACTTCGATGGTCGTCCTACAA 239 | no |
| Cluster-16493.40057 | LINE-1 retrotransposable element ORF2 protein | ptc-miR396e-3p | miRNA: 1 CTCAAGAAAGCTGTGGGAGA 20 * : *|| |||| |||||| : | | | | Target: 2923 CGCTTCTTTCGACACTCTCT 2904 | no |
| Cluster-16493.31604 | PREDICTED: coatomer subunit alpha-1-like isoform X1 | ptc-miR482a.1 | miRNA: 1 CCTACTCCTCCCATTCC 17 ||||* | |* | || || |: || Target: 1901 GGATAAGAAGGGTAGGG 1885 | no |
| Cluster-16493.26584 | Calcium-transporting ATPase 4 | ptc-miR482a.1 | miRNA: 1 CCTACTCCTCCCATTCC 17 *|| *| || || |* |||||| Target: 4880 TGAAGAGGAGTGTAAGG 4864 | no |
| Cluster-16493.41325 | Polyphenol oxidase, chloroplastic | ptc-miR1444d | miRNA: 1 CGAACGTTGACCGAATGT-GAA 21 || ||||| : |||||||||| * | * | Target: 85 GCTTGCAGCTGGCTTACACCCT 64 | yes |
| Cluster-16493.29872 | Basic leucine zipper 63 | ptc-miR172a | miRNA: 1 AGAATCTTGATGATGCTGCAT 21 | : || | |||||| |* |||| |* * | Target: 1241 TTTTAGAACTACGACGACCAA 1221 | yes |
| Cluster-16493.41106 | Putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 2 | ptc-miR172a | miRNA: 1 AGAATCTTGATGATGCTGCAT 21 | || || |||| | |*|*| || * ||| Target: 3710 TCTTAGAACTAGTCCGA-GTA 3691 | no |
| Cluster-16493.44708 | Heat shock protein 90-1 | ptc-miR319i | miRNA: 1 TTGGGCTGAAGGGAGCTCCC 20 ||| * ||: | | || : || || ||| * Target: 890 AACGCGGCTTCTCTCGAGGT 871 | yes |
| Cluster-16493.49274 | 18.2 kDa class I heat shock family protein | ptc-miR7812 | miRNA: 1 CTGTTATGAATTGATGGAGTG 21 * ||* ||| |||| || |||||*|| Target: 305 AACTATACTTAACTACCT-AC 286 | yes |
| Cluster-16493.45983 | endo-glucanase 2 family protein | ptc-miR6462e | miRNA: 1 TCTTATGCGTTTTTGTCTCT 20 |||| || ||| ||| : : : | |* *| Target: 590 AGAATACGCAAAGGTAG-TA 572 | yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, B.; Kang, Y.; Leng, J.; Xu, Q. Genome-Wide Analysis of the miRNA–mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra. Genes 2019, 10, 430. https://doi.org/10.3390/genes10060430
Zhou B, Kang Y, Leng J, Xu Q. Genome-Wide Analysis of the miRNA–mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra. Genes. 2019; 10(6):430. https://doi.org/10.3390/genes10060430
Chicago/Turabian StyleZhou, Bo, Yutong Kang, Jingtong Leng, and Qijiang Xu. 2019. "Genome-Wide Analysis of the miRNA–mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra" Genes 10, no. 6: 430. https://doi.org/10.3390/genes10060430
APA StyleZhou, B., Kang, Y., Leng, J., & Xu, Q. (2019). Genome-Wide Analysis of the miRNA–mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra. Genes, 10(6), 430. https://doi.org/10.3390/genes10060430




