Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Putative Chitinase Genes
2.2. Multiple Alignment and Phylogenetic Analysis
2.3. Identification of Conserved Domains
2.4. Visualization of Chitinase Genes on Chromosomes
2.5. Prediction of Cis-Acting Elements in Promoter Regions of Chitinase Genes
2.6. Signal Peptide Prediction
2.7. Plant Materials and Stress Treatments
2.8. qRT-PCR Analysis of the Chitinase Genes in A. nanus
2.9. Statistics
3. Results
3.1. Genome-wide Identification of chitinase Genes in A. nanus
3.2. Multiple Alignment and Phylogenetic Analysis
3.3. Conserved Domains
3.4. Intron-Exon Architecture
3.5. Prediction of the Cis-Acting Elements in Promoter Region of the Chitinase in A. nanus
3.6. Expression Levels of A. nanus Chitinases in Leaf, Stem, and Root
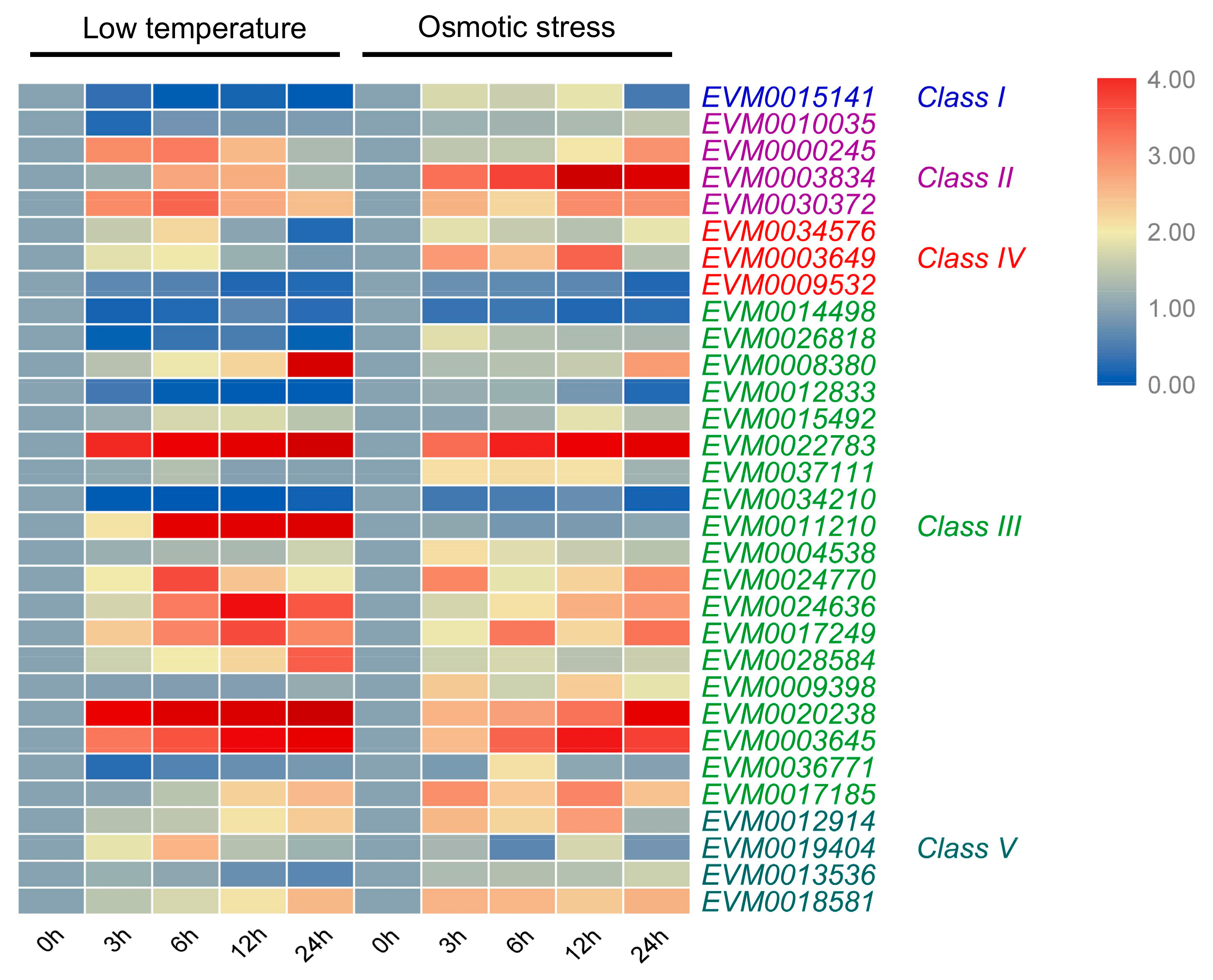
3.7. Expression Pattern of A. nanus Chitinases under Low Temperature Stress
3.8. Expression Pattern of A. nanus Chitinases under Osmotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Grover, A. Plant chitinases: Genetic diversity and physiological roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Rathore, J.S.; Ghosh, C. Pathogen-Associated Molecular Patterns and Their Perception in Plants. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 79–113. [Google Scholar]
- Newman, M.A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Zhang, S.; Stacey, G. Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 2004, 5, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Legrand, M.; Kauffmann, S.; Geoffroy, P.; Fritig, B. Biological function of pathogenesis-related proteins: Four tobacco pathogenesis-related proteins are chitinases. Proc. Natl. Acad. Sci. USA 1987, 84, 6750–6754. [Google Scholar] [CrossRef] [Green Version]
- Hengel, A.J.; van Kammen, A.; de Vries, S.C. A relationship between seed development, arabinogalactan-protein (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol. Plant. 2002, 114, 637–644. [Google Scholar] [CrossRef]
- Dyachok, J.V.; Wiweger, M.; Kenne, L.; von Arnold, S. Endogenous Nod-factor-like signal molecules promote early somatic embryo development in Norway spruce. Plant Physiol. 2002, 128, 523–533. [Google Scholar] [CrossRef]
- Kasprzewska, A.N.N.A. Plant chitinases-regulation and function. Cell. Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar]
- Zhong, R.; Kays, S.J.; Schroeder, B.P.; Ye, Z.H. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 2002, 14, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Malolepszy, A.; Kelly, S.; Sørensen, K.K.; James, E.K.; Kalisch, C.; Bozsoki, Z.; Panting, M.; Andersen, S.U.; Sato, S.; Tao, K.; et al. A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. Elife 2018, 7, e38874. [Google Scholar] [CrossRef] [PubMed]
- Kremer, N.; Philipp, E.E.; Carpentier, M.C.; Brennan, C.A.; Kraemer, L.; Altura, M.A.; Augustin, R.; Häsler, R.; Heath-Heckman, E.A.; Peyer, S.M.; et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 2013, 14, 183–194. [Google Scholar] [CrossRef]
- Liu, B.; Xue, X.; Cui, S.; Zhang, X.; Han, Q.; Zhu, L.; Kang, Z. Cloning and characterization of a wheat β-1, 3-glucanase gene induced by the stripe rust pathogen Puccinia striiformis f. sp. tritici. Mol. Biol. Rep. 2010, 37, 1045. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Ala, P.; Yang, D.S.; Hon, W.C.; Moffatt, B.A. Antifreeze protein produced endogenously in winter rye leaves. Plant Physiol. 1992, 100, 593–596. [Google Scholar] [CrossRef]
- Su, Y.; Xu, L.; Fu, Z.; Yang, Y.; Guo, J.; Wang, S.; Que, Y. ScChi, encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int. J. Mol. Sci. 2014, 15, 2738–2760. [Google Scholar] [CrossRef]
- Nakamura, T.; Ishikawa, M.; Nakatani, H.; Oda, A. Characterization of cold-responsive extracellular chitinase in bromegrass cell cultures and its relationship to antifreeze activity. Plant Physiol. 2008, 147, 391–401. [Google Scholar] [CrossRef]
- Jarząbek, M.; Pukacki, P.M.; Nuc, K. Cold-regulated proteins with potent antifreeze and cryoprotective activities in spruces (Picea spp.). Cryobiology 2009, 58, 268–274. [Google Scholar]
- Zhang, S.H.; Wei, Y.; Liu, J.L.; Yu, H.M.; Yin, J.H.; Pan, H.Y.; Baldwin, T.C. An apoplastic chitinase CpCHT1 isolated from the corolla of wintersweet exhibits both antifreeze and antifungal activities. Biol. Plant. 2011, 55, 141–148. [Google Scholar] [CrossRef]
- Jin Kim, Y.; Kook Hwang, B. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 2000, 108, 51–60. [Google Scholar] [CrossRef]
- Wu, Q.; Gao, S.; Pan, Y.B.; Su, Y.; Grisham, M.P.; Guo, J.; Que, Y. Heterologous expression of a Glyoxalase I gene from sugarcane confers tolerance to several environmental stresses in bacteria. PeerJ 2018, 6, e5873. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, P.; Deswal, R. A novel class I Chitinase from Hippophae rhamnoides: Indications for participating in ICE-CBF cold stress signaling pathway. Plant Sci. 2017, 259, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, P.A.; de Vries, S.C. Arabidopsis chitinases: A genomic survey. Arab. Book/Am. Soc. Plant Biol. 2002, 1, e0023. [Google Scholar]
- Tobias, P.A.; Christie, N.; Naidoo, S.; Guest, D.I.; Külheim, C. Identification of the Eucalyptus grandis chitinase gene family and expression characterization under different biotic stress challenges. Tree Physiol. 2017, 37, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Huang, R.F.; Song, J.L.; Huang, M.R.; Xu, L.A. Genomewide analysis of the chitinase gene family in Populus trichocarpa. J. Genet. 2013, 92, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Tan, X. Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B. Molecular evolution and functional divergence of chitinase gene family in Hevea brasiliensis genome. Winnower 2015, 6. [Google Scholar] [CrossRef]
- Mir, Z.A.; Ali, S.; Shivaraj, S.M.; Bhat, J.A.; Singh, A.; Yadav, P.; Rawat, S.; Paplao, P.K.; Grover, A. Genome-wide identification and characterization of Chitinase gene family in Brassica juncea and Camelina sativa in response to Alternaria brassicae. Genomics 2019. [Google Scholar] [CrossRef]
- Su, Y.; Xu, L.; Wang, S.; Wang, Z.; Yang, Y.; Chen, Y.; Que, Y. Identification, phylogeny, and transcript of chitinase family genes in sugarcane. Sci. Rep. 2015, 5, 10708. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Tian, L.; Wang, G.; Zhang, X.; Wang, X.; Guo, W. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 2016, 6, 29022. [Google Scholar] [CrossRef]
- Gao, F.; Li, H.; Xiao, Z.; Wei, C.; Feng, J.; Zhou, Y. De novo transcriptome analysis of Ammopiptanthus nanus and its comparative analysis with A. mongolicus. Trees 2018, 32, 287–300. [Google Scholar] [CrossRef]
- Gao, F.; Wang, X.; Li, X.; Xu, M.; Li, H.; Abla, M.; Sun, H.; Wei, S.; Feng, J.; Zhou, Y. Long-read sequencing and de novo genome assembly of Ammopiptanthus Nanus, a desert shrub. GigaScience 2018, 7, giy074. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Academic Press: Waltham, MA, USA, 1965; pp. 97–166. [Google Scholar]
- Al Ait, L.; Yamak, Z.; Morgenstern, B. DIALIGN at GOBICS—multiple sequence alignment using various sources of external information. Nucleic Acids Res. 2013, 41, W3–W7. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. Gerf. Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, J.; Wei, S.; Li, Z.; Wang, N.; Li, H.; Zhang, F. Transcriptomic analysis of drought stress responses in Ammopiptanthus mongolicus leaves using the RNA-Seq technique. PLoS ONE 2015, 10, e0124382. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta DeltaC (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Neuhaus, J.M.; Fritig, B.; Linthorst, H.J.M.; Meins, F.; Mikkelsen, J.D.; Ryals, J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996, 14, 102–104. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad1, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef]
- Hamel, F.; Boivin, R.; Tremblay, C.; Bellemare, G. Structural and evolutionary relationships among chitinases of flowering plants. J. Mol. Evol. 1997, 44, 614–624. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Iu, B.; Singh, J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 1996, 30, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, T.; Shinshi, H.; Suzuki, K. Rapid and transient activation of transcription of the ERF3 Gene by wounding in tobacco leaves possible involvement of NtWRKYs and autorepression. J. Biol. Chem. 2004, 279, 55355–55361. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000, 24, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Zhou, X.Y.; Wang, Y.G.; Zhou, S.F.; Fu, F.L.; Li, W.C. A betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus enhances tolerance of Arabidopsis to high salt and drought stresses. Plant Growth Regul. 2017, 83, 265–276. [Google Scholar] [CrossRef]
- Yu, H.Q.; Han, N.; Zhang, Y.Y.; Tao, Y.; Chen, L.; Liu, Y.P.; Li, W.C. Cloning and characterization of vacuolar H+-pyrophosphatase gene (AnVP1) from Ammopiptanthus nanus and its heterologous expression enhances osmotic tolerance in yeast and Arabidopsis thaliana. Plant Growth Regul. 2017, 81, 385–397. [Google Scholar] [CrossRef]
- Jekel, P.A.; Hartmann, J.B.H.; Beintema, J.J. The primary structure of hevamine, an enzyme with lysozyme/chitinase activity from Hevea brasiliensis latex. Eur. J. Biochem. 1991, 200, 123–130. [Google Scholar] [CrossRef]
- Graça, I.; Liang, J.; Guilherme, M.; Tavares, P.; Ferreira-Pinto, M.M.; Melo, A.M.; Ribeiro-Barros, A.I.; Pereira, A.S. Cloning, overexpression and functional characterization of a class III chitinase from Casuarina glauca nodules. Symbiosis 2016, 70, 139–148. [Google Scholar] [CrossRef]
- Liu, X.; Grabherr, H.M.; Willmann, R.; Kolb, D.; Brunner, F.; Bertsche, U.; Kühner, D.; Franz-Wachtel, M.; Amin, B.; Felix, G.; et al. Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. Elife 2014, 3, e01990. [Google Scholar] [CrossRef]
- Yeboah, N.A.; Arahira, M.; Zhang, D.; Kadokura, K.; Watanabe, A.; Fukazawa, C. A class III acidic endochitinase is specifically expressed in the developing seeds of soybean (Glycine max [L.] Merr.). Plant Mol. Biol 1998, 36, 407–415. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. J. Nat. Rev. Genet. 2006, 7, 211. [Google Scholar]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Mourier, T.; Penny, D. The biology of intron gain and loss. Trends Genet. 2006, 22, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef]
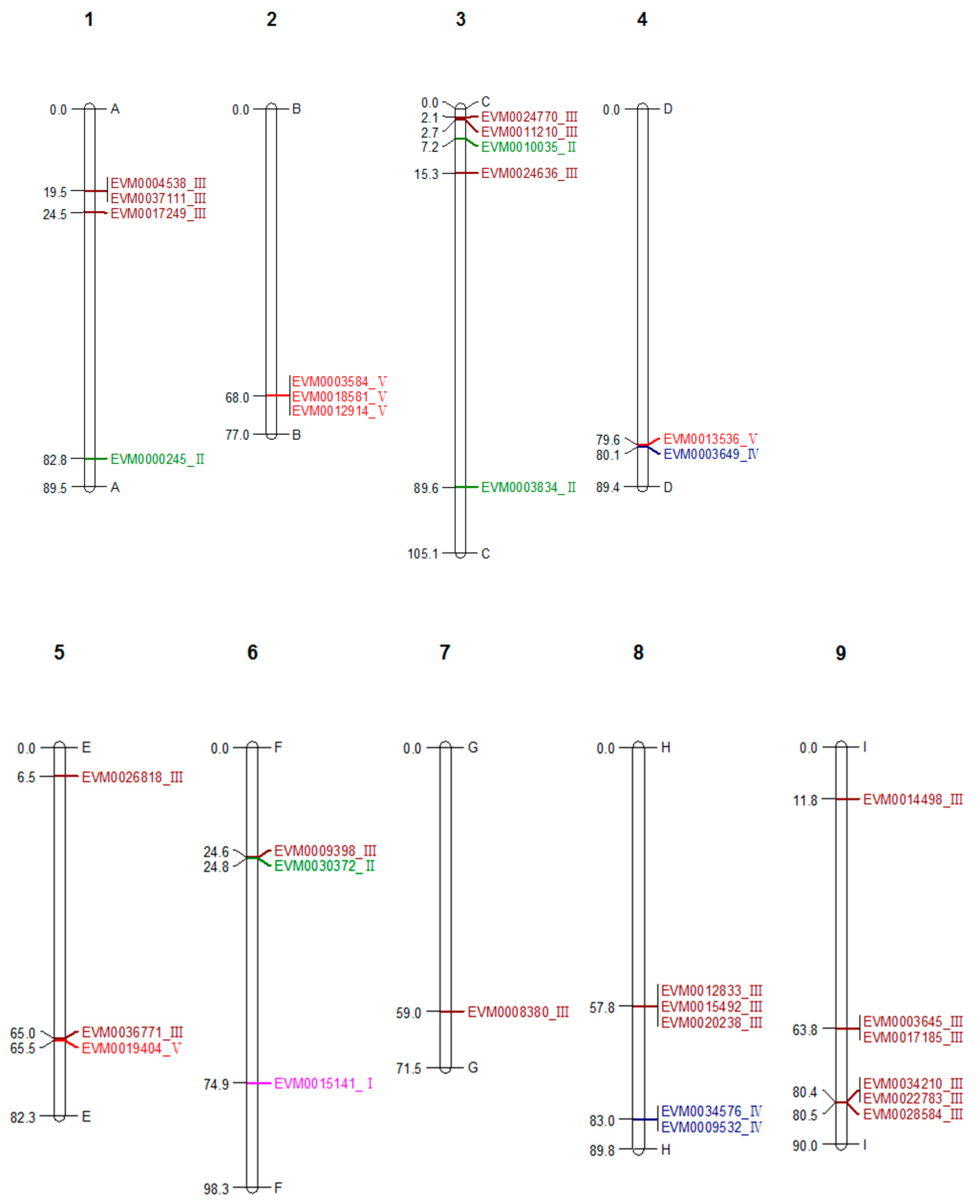
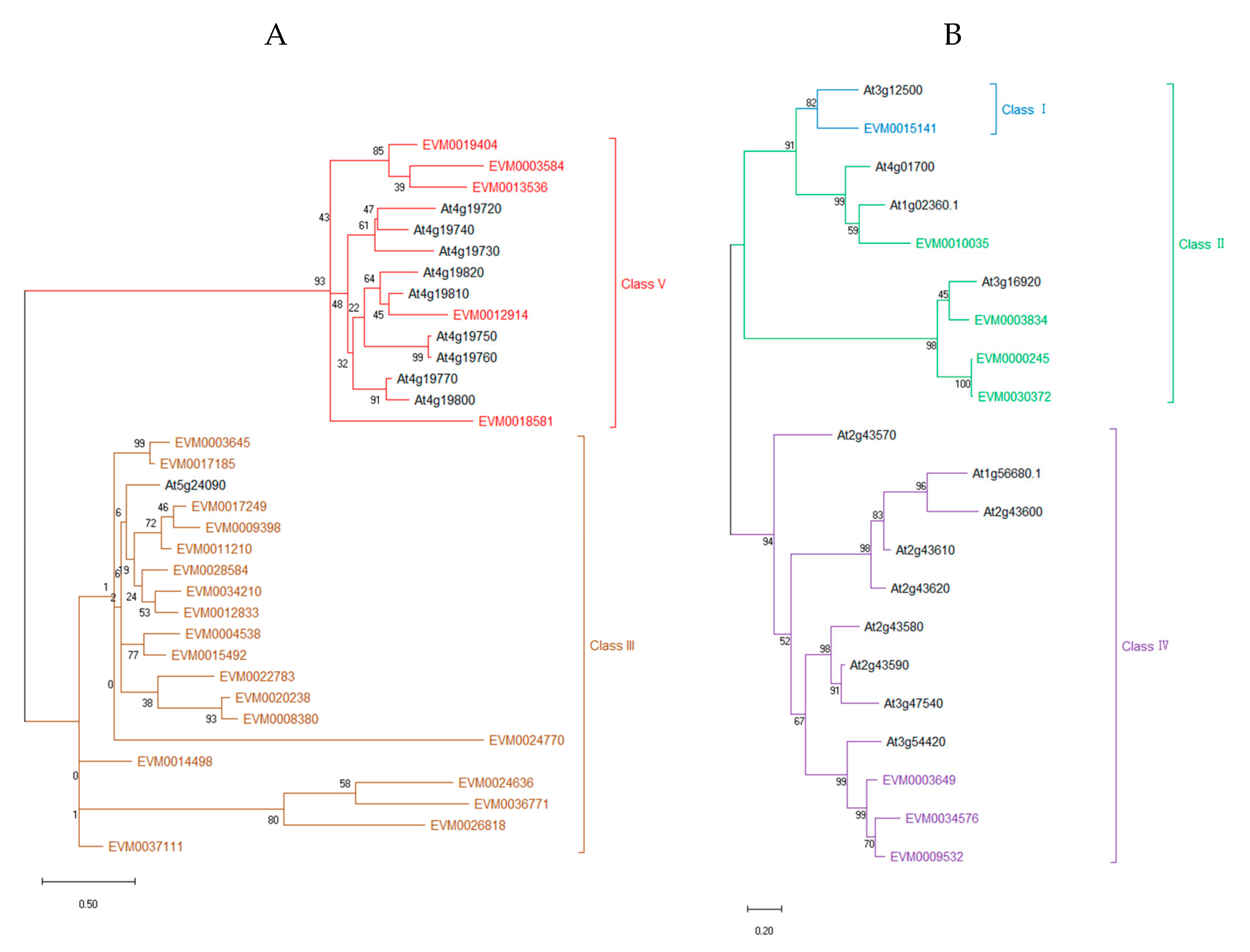


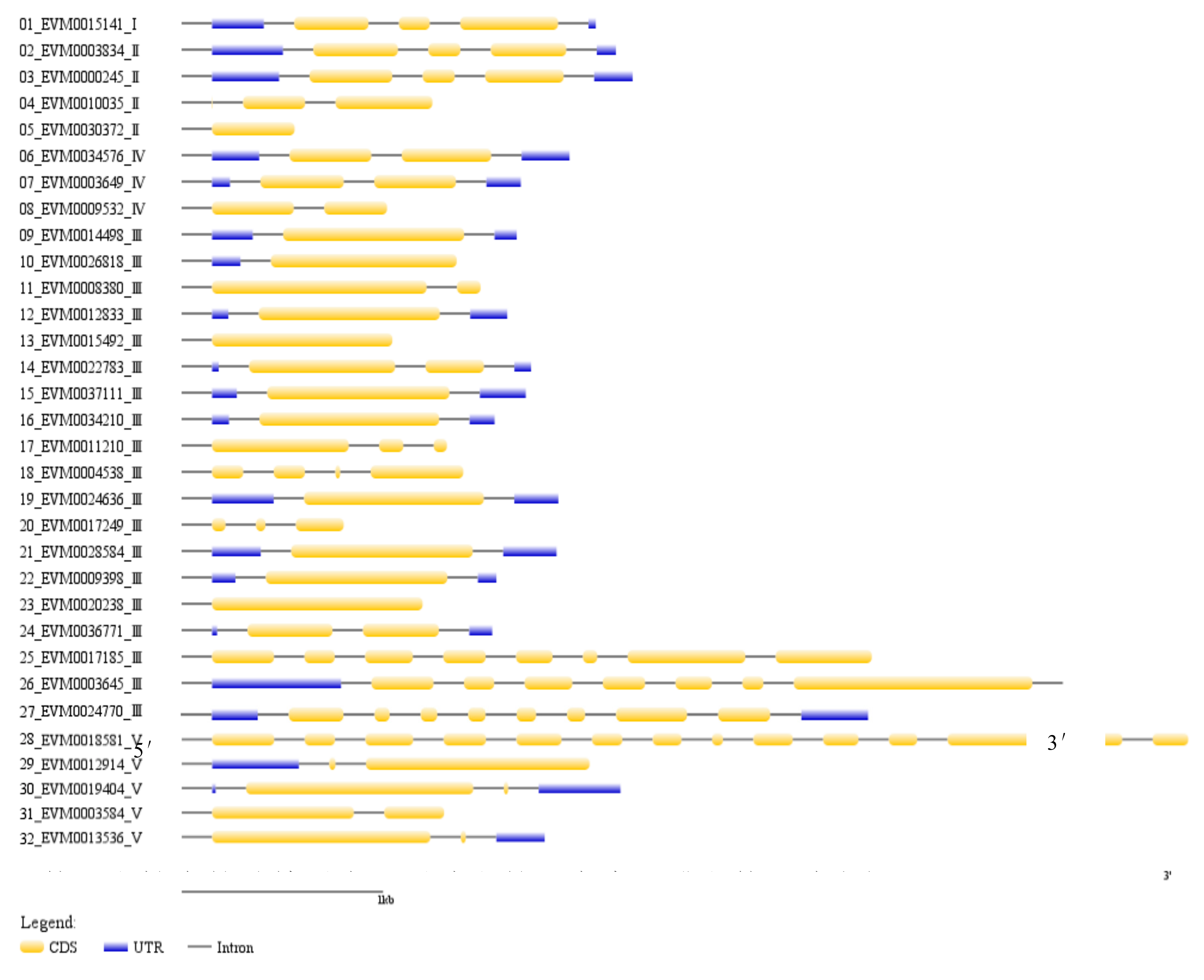

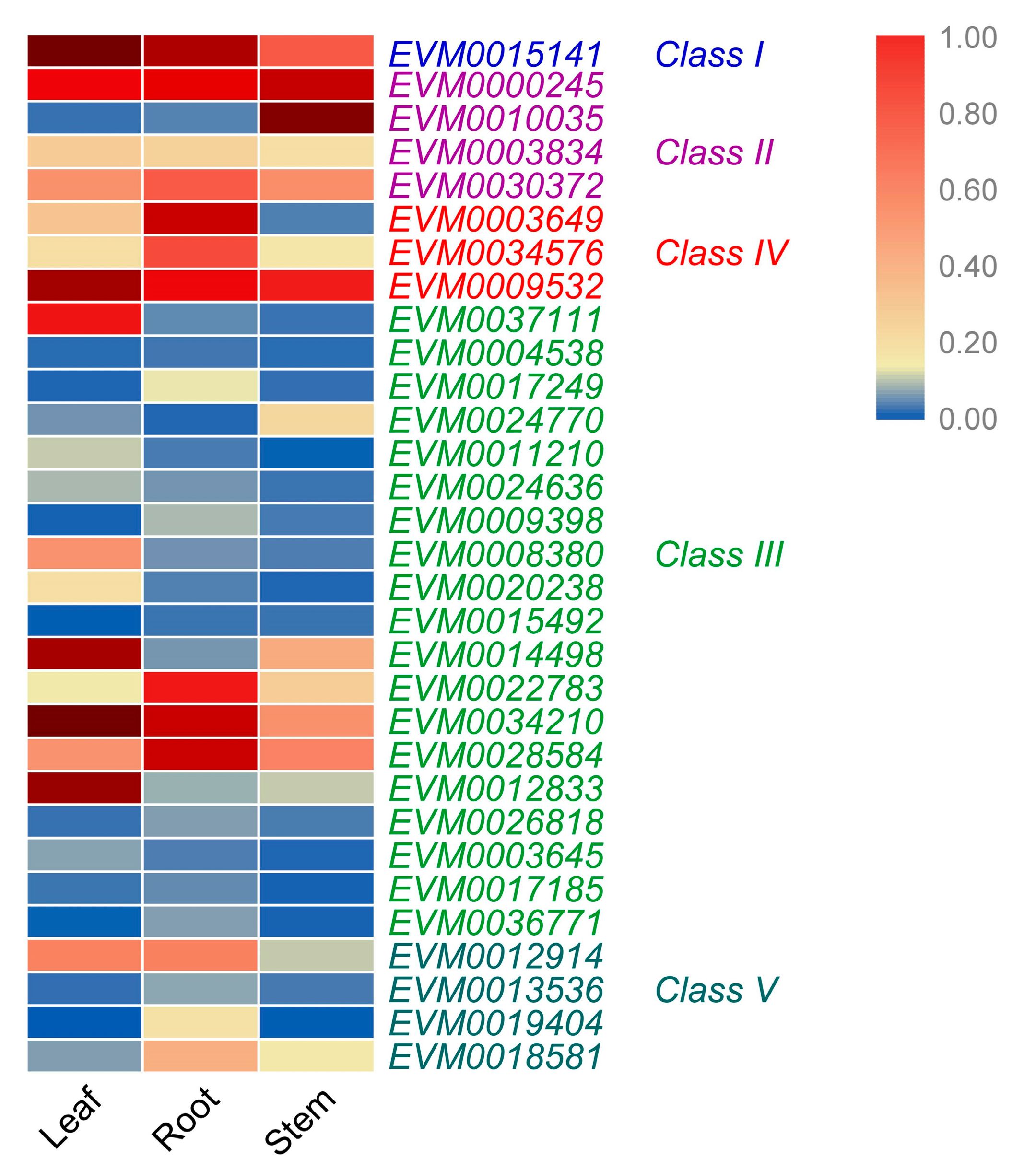
| Sequence ID | Genome Location | Domains | Class | Amino Acid Length | Signal Peptide | Number of Introns | Chitin Binding Region | Predicted pI | Predicted Mw |
|---|---|---|---|---|---|---|---|---|---|
| EVM0015141 | ch06:74872028--74873992 | GH19 | I | 344 | 1-24 | 4 | 50--69 | 8.42 | 36,164.62 |
| EVM0000245 | ch01:82754230--82758136 | GH19 | II | 328 | 1-24 | 4 | NO | 5.82 | 35,570.39 |
| EVM0010035 | ch03:67952710--67953891 | GH19 | II | 270 | 1-17 | 2 | NO | 5.81 | 29,087.94 |
| EVM0003834 | ch03:89647173--89650518 | GH19 | II | 326 | 1-21 | 4 | NO | 7.48 | 34,959.76 |
| EVM0030372 | ch06:24817881--24818291 | GH19 | II | 139 | 1-29 | 0 | NO | 8.58 | 15,484.92 |
| EVM0003649 | ch04:80079800--80081378 | GH19 | IV | 279 | 1-26 | 3 | 31--50 | 4.70 | 29,787.24 |
| EVM0034576 | ch08:83000063--83004274 | GH19 | IV | 291 | NO | 3 | 40--64 | 4.22 | 30,666.83 |
| EVM0009532 | ch08:83008232--83009062 | GH19 | IV | 245 | NO | 1 | NO | 4.88 | 26,440.78 |
| EVM0037111 | ch01:19478187--19479445 | GH18 | III | 309 | 1-27 | 2 | NO | 6.43 | 32,622.71 |
| EVM0004538 | ch01:19470407--19471892 | GH18 | III | 271 | 1-23 | 3 | NO | 5.83 | 28,923.78 |
| EVM0017249 | ch01:24538090--24543941 | GH18 | III | 119 | NO | 2 | NO | 5.10 | 12,987.12 |
| EVM0024770 | ch03:2089454--2095600 | GH18 | III | 441 | 1-20 | 9 | NO | 8.59 | 49,055.54 |
| EVM0011210 | ch03:2653696--2655594 | GH18 | III | 296 | 1-28 | 2 | NO | 4.87 | 31,084.80 |
| EVM0024636 | ch03:15250701--15252121 | GH18 | III | 305 | 1-29 | 2 | NO | 5.27 | 33,518.83 |
| EVM0009398 | ch06:24575731--24576843 | GH18 | III | 308 | 1-25 | 2 | NO | 6.80 | 31,905.08 |
| EVM0008380 | ch07:59025620--59027551 | GH18 | III | 404 | 1-27 | 1 | NO | 5.46 | 43,543.43 |
| EVM0020238 | ch07:57782685--57783731 | GH18 | III | 356 | 1-30 | 0 | NO | 4.56 | 38,587.45 |
| EVM0015492 | ch08:57768990--57769886 | GH18 | III | 306 | 1-23 | 0 | NO | 8.92 | 32,347.84 |
| EVM0014498 | ch09:11798331--11799544 | GH18 | III | 307 | 1-28 | 2 | NO | 8.97 | 32,399.49 |
| EVM0022783 | ch09:80432948--80446622 | GH18 | III | 346 | 1-27 | 3 | NO | 5.17 | 37,050.17 |
| EVM0034210 | ch09:80420661--80421764 | GH18 | III | 305 | 1-24 | 2 | NO | 5.04 | 31,361.14 |
| EVM0028584 | ch09:80454721--80456132 | GH18 | III | 308 | 1-22 | 2 | NO | 8.98 | 32,352.94 |
| EVM0012833 | ch08:57760416--57761582 | GH18 | III | 307 | 1-25 | 2 | NO | 4.44 | 31,602.09 |
| EVM0026818 | ch05:6506791--6507856 | GH18 | III | 315 | 1-29 | 1 | NO | 5.56 | 33,909.98 |
| EVM0003645 | ch08:63789032--63793200 | GH18 | III | 804 | 1-29 | 8 | NO | 8.35 | 88,243.97 |
| EVM0017185 | ch08:63796631--63800128 | GH18 | III | 751 | 1-31 | 7 | NO | 8.29 | 82,885.73 |
| EVM0036771 | ch04:6503824--6505708 | GH18 | III | 269 | NO | 3 | NO | 5.77 | 30,665.56 |
| EVM0012914 | ch02:67985626--67991461 | GH18 | V | 390 | 1-20 | 2 | NO | 8.63 | 41,046.00 |
| EVM0003584 | ch02:67952710--67953891 | GH18 | V | 341 | 1-24 | 1 | NO | 6.81 | 36,866.35 |
| EVM0013536 | ch04:79575721--79578939 | GH18 | V | 379 | 1-21 | 2 | NO | 9.12 | 40,742.48 |
| EVM0019404 | ch05:65508611--65510368 | GH18 | V | 393 | 1-21 | 3 | NO | 7.78 | 43,121.80 |
| EVM0018581 | ch01:67959345--67975423 | GH18 | V | 1027 | 1-22 | 12 | NO | 7.25 | 114,928.43 |
| A. nanus | A. thaliana | B. rapa | P. trichocarpa | E. grandis | G. raimondii | H. brasiliensis | B. juncea | C. sativa | |
|---|---|---|---|---|---|---|---|---|---|
| Class I | 1 | 1 | 5 | 11 | 1 | 5 | 7 | 10 | 4 |
| Class II | 4 | 2 | 5 | 3 | 6 | 5 | 1 | 6 | 21 |
| Class III | 19 | 1 | 2 | 13 | 10 | 10 | 16 | 2 | 3 |
| Class IV | 3 | 9 | 17 | 5 | 14 | 7 | 5 | 25 | 26 |
| Class V | 5 | 9 | 4 | 5 | 26 | 19 | 10 | 4 | 25 |
| Total | 32 | 24 | 33 | 37 | 67 | 47* | 39 | 47 | 79 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, S.; Wang, Y.; Li, Z.; Shi, W.; Gao, F.; Zhou, Y.; Zhang, G.; Feng, J. Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus. Genes 2019, 10, 472. https://doi.org/10.3390/genes10060472
Cao S, Wang Y, Li Z, Shi W, Gao F, Zhou Y, Zhang G, Feng J. Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus. Genes. 2019; 10(6):472. https://doi.org/10.3390/genes10060472
Chicago/Turabian StyleCao, Shilin, Ying Wang, Zhiqiang Li, Wei Shi, Fei Gao, Yijun Zhou, Genfa Zhang, and Jinchao Feng. 2019. "Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus" Genes 10, no. 6: 472. https://doi.org/10.3390/genes10060472
APA StyleCao, S., Wang, Y., Li, Z., Shi, W., Gao, F., Zhou, Y., Zhang, G., & Feng, J. (2019). Genome-Wide Identification and Expression Analyses of the Chitinases under Cold and Osmotic Stress in Ammopiptanthus nanus. Genes, 10(6), 472. https://doi.org/10.3390/genes10060472





