Screening the Reference Genes for Quantitative Gene Expression by RT-qPCR During SE Initial Dedifferentiation in Four Gossypium hirsutum Cultivars that Have Different SE Capability
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Selection of Reference Genes and Design of Primers
2.4. RT-qPCR Analysis
2.5. Data Analysis
2.6. Validation of Reference Genes
3. Results
3.1. Isolation of Candidate Reference Genes in Different G. hirsutum Cultivars
3.2. Verification of Primer Specificity and PCR Amplification Efficiency
3.3. Expression Profile Analysis of the Candidate Reference Genes at Different Induction Stages in Different Cotton Cultivars
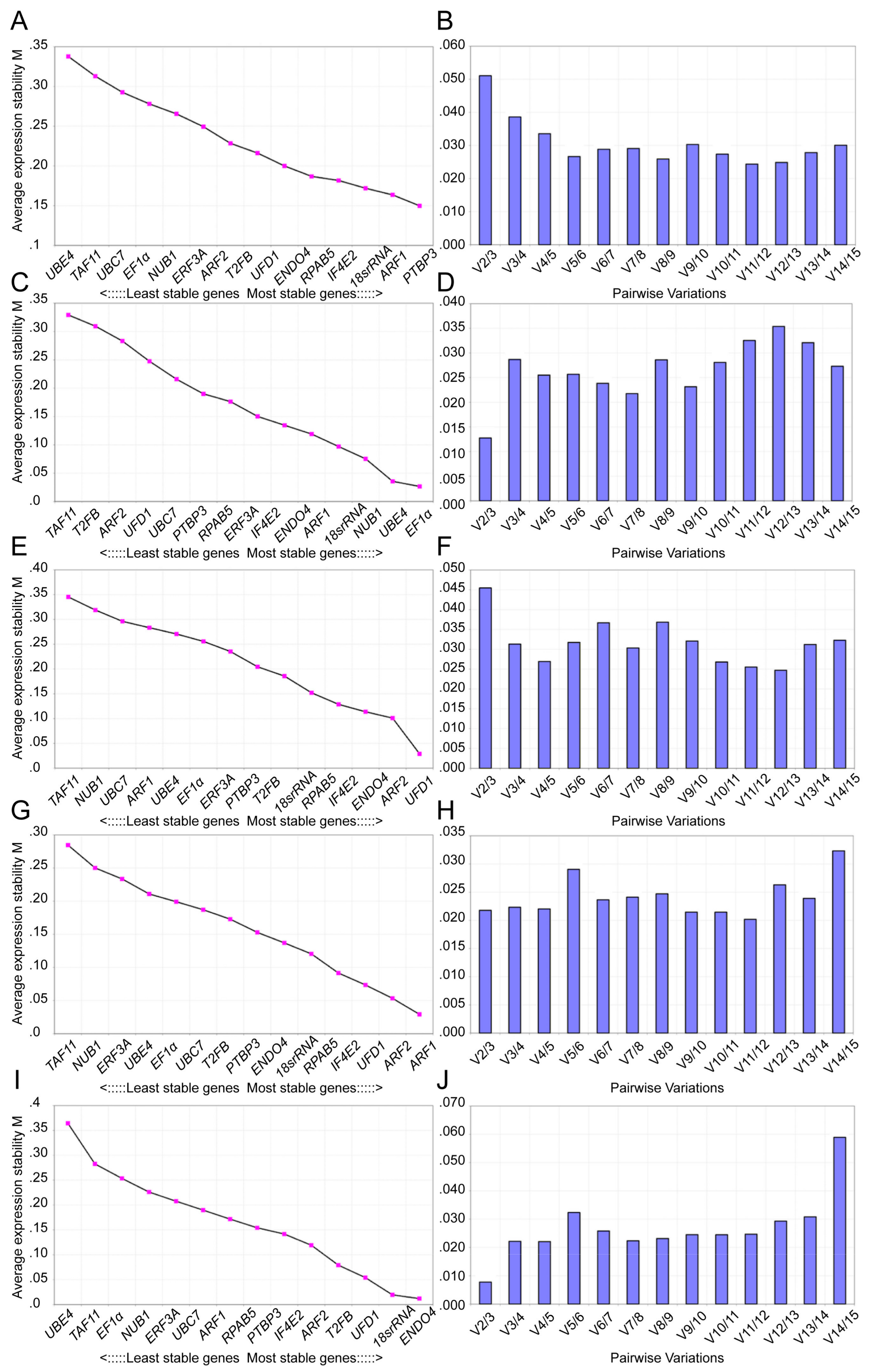
3.4. Expression Stability Analysis of Reference Genes
3.5. Expression Stability Validation of the Reference Genes of 18S rRNA and ENDO4
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, C.; Liu, Q.; Zhang, Z.; Zheng, B.; Bao, M. De novo comparative transcriptome analysis provides new insights into sucrose induced somatic embryogenesis in camphor tree (Cinnamomum camphora L.). BMC Genom. 2016, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference Gene Selection for Quantitative Real-time PCR Normalization in Caragana intermedia under Different Abiotic Stress Conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Hong, S.Y.; Seo, P.J.; Yang, M.S.; Xiang, F.; Park, C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Zhang, S.; Zeng, Y.; Yi, X.; Zhang, Y. Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Sci. Rep. 2016, 6, 30363. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, H.; Zhang, R.; Yang, Q.; Liu, Y.; Zang, X. Selection of reference gene from Gracilaria lemaneiformis under temperature stress. J. Appl. Phycol. 2014, 27, 1–8. [Google Scholar] [CrossRef]
- Sun, H.P.; Li, F.; Ruan, Q.M.; Zhong, X.H. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. Plant Physiol. Biochem. 2016, 108, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi-Darvari, F.; Noor, N.M.; Ismanizan, I. Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell Tissue Organ Cult. 2015, 120, 407–422. [Google Scholar] [CrossRef]
- Kumria, R.; Sunnichan, V.G.; Das, D.K.; Gupta, S.K.; Reddy, V.S.; Bhatnagar, R.K.; Leelavathi, S. High-frequency somatic embryo production and maturation into normal plants in cotton (Gossypium hirsutum) through metabolic stress. Plant Cell Rep. 2003, 21, 635–639. [Google Scholar] [PubMed]
- Yang, X.; Zhang, X.; Yuan, D.; Jin, F.; Zhang, Y.; Jiao, X. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012, 12, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Zhu, H.G.; Tian, W.G.; Zhu, S.H.; Xiong, X.P.; Sun, Y.Q.; Zhu, Q.H.; Sun, J. De novo transcriptome analysis reveals insights into dynamic homeostasis regulation of somatic embryogenesis in upland cotton (G. hirsutum L.). Plant Mol. Biol. 2016, 92, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Zhang, X.L. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Fausto, A.K.S.; Silva, T.D.F.; Romanel, E.; Vaslin, M.F.S. microRNAs as reference genes for quantitative PCR in cotton. PLoS ONE 2017, 12, e0174722. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.Y.; Guo, X.; Liang, S.; Zhu, H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 2006, 50, 519–524. [Google Scholar] [CrossRef]
- Cao, A.; Zheng, Y.; Yu, Y.; Wang, X.; Shao, D.; Sun, J.; Cui, B. Comparative Transcriptome Analysis of SE initial dedifferentiation in cotton of different SE capability. Sci. Rep. 2017, 7, 8583. [Google Scholar] [CrossRef] [PubMed]
- Jonge, H.; Fehrmann, R.; Bont, E.; Hofstra, R.; Gerbens, F.; Kamps, W.; Vries, E.; Zee, A.; Meerman, G.; Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, B.; Tan, Z.; Liu, J.; Yang, Z.; Li, Z.; Huang, B. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015, 34, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yan, W.; Huang, X.; Jiang, L.; Wang, H.; Li, J. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 2015, 15, 439. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–153. [Google Scholar] [CrossRef]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef]
- Xue, C.; Yingji, M.; Shengwei, H.; Jun, N.; Weili, L.; Jinyan, H. Selection of suitable reference genes for quantitative real-time PCR in Sapium sebiferum. Front. Plant Sci. 2017, 8, 637. [Google Scholar]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene Selection for RT-qPCR Analysis of Flower Development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 11, 287. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Sci. 2009, 176, 702–706. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of Reliable Reference Genes for Gene Expression Studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 18, 1547. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Qi, J.; Zhang, G.; Xu, J.; Tao, A.; Fang, P.; Su, J. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 2015, 14, 848. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.; Shi, S.; Liu, J.; Cheng, T.; Xue, L.; Yang, X.; Yang, W.; Lan, Q.; Jiang, Z. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PCR. PLoS ONE 2012, 7, e33278. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.M.; Sá, P.H.; Castro, T.L.; Carvalho, R.D.; Pinto, A.; Gil, D.J.P.; Silva, A. Reference genes for RT-qPCR studies in Corynebacterium pseudotuberculosisi dentified through analysis of RNA-seq data. Antonie Leeuwenhoek 2014, 106, 605–614. [Google Scholar] [CrossRef]




| Gene Name | Accession Number | Primer Sequence (5′-3′) | Product Size (bp) | Ea (%) | R2 |
|---|---|---|---|---|---|
| 18S rRNA | XM_016849259 | TTACGCAATGCGCTCTGGA | 117 | 104.70 | 0.9968 |
| ACCGCAGAGCTGACAGATG | |||||
| ARF1 | XM_016856733 | CTGTGAGCAGAAAGTGGAAAGC | 111 | 104.61 | 0.9996 |
| CAGCTGCATCAAGACCCACC | |||||
| ARF2 | XM_016840408 | CCACTTCTGGTGAAGGTCTGT | 118 | 100.33 | 0.9980 |
| AACTCTAAAAGGGGCCAGCA | |||||
| EF1α | XM_016892582 | CAGCTTCAGATCGCTTCTATTTCT | 124 | 100.07 | 0.9998 |
| TGGCCAGTGGTGGTTGACTT | |||||
| ENDO4 | XM_016854965. | TTGACAGAGGCGCTGATGTT | 116 | 100.44 | 0.9905 |
| CTGCGGTACCAACTGACTGT | |||||
| ERF3A | XM_016815421 | GCCCTATTTGCCACAAAACCC | 140 | 101.56 | 0.9886 |
| TTCAGGAATGAGCGTGGCAT | |||||
| IF4E2 | XM_016823302 | CAAGACTGCAACGAATGAGGC | 144 | 100.36 | 0.9894 |
| GCTCAAACATTGTATCGACCTTTCA | |||||
| NUB1 | XM_016834127 | TTGCACTACATATGAGGTTGGAGTT | 132 | 101.60 | 0.9677 |
| AGGCTTCATCAGGCACTTGTA | |||||
| PTBP3 | XM_016838300. | GTCCTTGCAAATGGCGGAAG | 140 | 102.08 | 0.9961 |
| CCTGATTCTTTGCACGGAGC | |||||
| RPAB5 | XM_016880466 | CTTCACCTGCGGAAAGGTCA | 113 | 99.85 | 1.0643 |
| AGCAGTACCGAACCAATCCC | |||||
| T2FB | XM_016852332 | GGATCGCGGGGAATTGGAA | 149 | 99.09 | 1.0030 |
| TGCCTCTCTTATTGTACACGCA | |||||
| TAF11 | XM_016821843 | TCGTCTGCATTAGAGAGTCGC | 138 | 96.13 | 0.9999 |
| GGCTGTTCAGCTCATCCTCA | |||||
| UBC7 | XM_016864885 | GCTGGACGCCAGTACATACA | 144 | 99.21 | 1.0997 |
| GGCTGACCTTCCTCCTGAAT | |||||
| UBE4 | XM_016812715 | TGGGCCCCTTTTTCCATGTT | 109 | 120.30 | 0.9999 |
| TCAGCTGCTCGTCTAGTTGATG | |||||
| UFD1 | XM_016828251 | TGTCAGCCGTTCTAAGGAAACA | 107 | 115.58 | 0.9773 |
| ACTTTCTCCCGGTGAATGGC |
| Rank | Gene | all | Gene | YZ1 | Gene | R15 | Gene | X33 | Gene | X42 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 18S rRNA | 0.27 | ARF1 | 0.24 | ENDO4 | 0.25 | ARF2 | 0.21 | ARF2 | 0.26 |
| 2 | ENDO4 | 0.27 | 18S rRNA | 0.25 | IF4E2 | 0.26 | ARF1 | 0.21 | 18S rRNA | 0.26 |
| 3 | ARF1 | 0.29 | RPAB5 | 0.26 | 18S rRNA | 0.27 | ENDO4 | 0.21 | UFD1 | 0.26 |
| 4 | PTBP3 | 0.29 | ENDO4 | 0.26 | UFD1 | 0.29 | 18S rRNA | 0.22 | T2FB | 0.26 |
| 5 | RPAB5 | 0.30 | PTBP3 | 0.27 | RPAB5 | 0.30 | UFD1 | 0.23 | ENDO4 | 0.27 |
| 6 | IF4E2 | 0.30 | IF4E2 | 0.28 | ARF2 | 0.31 | PTBP3 | 0.23 | ARF1 | 0.31 |
| 7 | UFD1 | 0.30 | NUB1 | 0.29 | PTBP3 | 0.34 | UBE4 | 0.27 | PTBP3 | 0.32 |
| 8 | ARF2 | 0.34 | UBE4 | 0.31 | UBE4 | 0.35 | EF1α | 0.27 | ERF3A | 0.32 |
| 9 | ERF3A | 0.34 | EF1α | 0.32 | ARF1 | 0.35 | IF4E2 | 0.27 | UBC7 | 0.33 |
| 10 | T2FB | 0.35 | UBC7 | 0.33 | ERF3A | 0.35 | RPAB5 | 0.28 | IF4E2 | 0.34 |
| 11 | EF1α | 0.35 | ERF3A | 0.38 | EF1α | 0.36 | UBC7 | 0.30 | RPAB5 | 0.36 |
| 12 | NUB1 | 0.38 | UFD1 | 0.40 | UBC7 | 0.37 | T2FB | 0.32 | NUB1 | 0.40 |
| 13 | UBC7 | 0.38 | ARF2 | 0.43 | T2FB | 0.38 | ERF3A | 0.36 | EF1α | 0.41 |
| 14 | TAF11 | 0.45 | T2FB | 0.45 | NUB1 | 0.48 | NUB1 | 0.37 | TAF11 | 0.47 |
| 15 | UBE4 | 0.50 | TAF11 | 0.46 | TAF11 | 0.52 | TAF11 | 0.51 | UBE4 | 0.90 |
| Rank | Gene | All | Gene | YZ1 | Gene | R15 | Gene | X33 | Gene | X42 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 18S rRNA | 0.08 | RPAB5 | 0.04 | ENDO4 | 0.01 | ENDO4 | 0.02 | ARF2 | 0.04 |
| 2 | ENDO4 | 0.09 | ARF1 | 0.05 | IF4E2 | 0.03 | ARF2 | 0.05 | T2FB | 0.06 |
| 3 | ARF1 | 0.10 | 18S rRNA | 0.06 | 18S rRNA | 0.05 | 18S rRNA | 0.05 | UFD1 | 0.08 |
| 4 | PTBP3 | 0.11 | PTBP3 | 0.08 | UFD1 | 0.13 | PTBP3 | 0.06 | 18S rRNA | 0.09 |
| 5 | IF4E2 | 0.13 | IF4E2 | 0.11 | RPAB5 | 0.13 | ARF1 | 0.06 | ENDO4 | 0.10 |
| 6 | RPAB5 | 0.13 | ENDO4 | 0.12 | ARF2 | 0.14 | UFD1 | 0.09 | ERF3A | 0.11 |
| 7 | UFD1 | 0.13 | UBC7 | 0.15 | UBE4 | 0.17 | EF1α | 0.11 | ARF1 | 0.12 |
| 8 | ARF2 | 0.17 | UBE4 | 0.17 | PTBP3 | 0.18 | IF4E2 | 0.14 | UBC7 | 0.14 |
| 9 | ERF3A | 0.17 | NUB1 | 0.17 | ARF1 | 0.19 | UBE4 | 0.15 | PTBP3 | 0.16 |
| 10 | EF1α | 0.18 | EF1α | 0.19 | ERF3A | 0.19 | UBC7 | 0.15 | EF1α | 0.20 |
| 11 | T2FB | 0.18 | UFD1 | 0.23 | EF1α | 0.20 | RPAB5 | 0.16 | IF4E2 | 0.20 |
| 12 | UBC7 | 0.21 | ERF3A | 0.23 | UBC7 | 0.20 | T2FB | 0.20 | RPAB5 | 0.22 |
| 13 | NUB1 | 0.21 | ARF2 | 0.27 | T2FB | 0.22 | ERF3A | 0.22 | NUB1 | 0.23 |
| 14 | TAF11 | 0.27 | T2FB | 0.28 | NUB1 | 0.32 | NUB1 | 0.23 | TAF11 | 0.25 |
| 15 | UBE4 | 0.31 | TAF11 | 0.28 | TAF11 | 0.35 | TAF11 | 0.35 | UBE4 | 0.60 |
| Total | YZ1 | R15 | X33 | X42 | |
|---|---|---|---|---|---|
| Gene (Ra) | TAF11 (0.94) | TAF11 (0.46) | TAF11 (0.95) | TAF11 (0.98) | TAF11 (0.52) |
| CVb ± SDc | 1.63 ± 0.35 | 0.53 ± 0.11 | 2.23 ± 0.48 | 1.81 ± 0.39 | 1.72 ± 0.36 |
| Gene (R) | UBC7 (0.94) | ARF2 (0.44) | UBC7 (1.00) | UBC7 (0.99) | UBE4 (0.98) |
| CV ± SD | 2.18 ± 0.45 | 0.62 ± 0.12 | 2.52 ± 0.53 | 2.94 ± 0.61 | 2.46 ± 0.48 |
| Gene (R) | T2FB (0.95) | T2FB (0.44) | ARF1 (1.00) | END04 (1.00) | UBC7 (0.95) |
| CV ± SD | 2.74 ± 0.59 | 0.75 ± 0.16 | 2.65 ± 0.50 | 3.23 ± 0.69 | 2.49 ± 0.51 |
| Gene (R) | PTBP3 (0.98) | UBC7 (0.94) | PTBP3 (1.00) | EF1α (0.99) | EF1α (0.93) |
| CV ± SD | 2.99 ± 0.57 | 0.94 ± 0.19 | 2.72 ± 0.52 | 3.29 ± 0.64 | 2.83 ± 0.54 |
| Gene (R) | 18S rRNA (0.98) | RPAB5 (0.98) | T2FB (0.96) | T2FB (0.97) | ARF1 (0.99) |
| CV ± SD | 3.09 ± 0.64 | 1.48 ± 0.28 | 2.91 ± 0.63 | 3.33 ± 0.73 | 3.05 ± 0.57 |
| Gene (R) | ENDO4 (0.99) | UFD1 (0.67) | 18S rRNA (0.99) | PTBP3 (1.00) | T2FB (1.00) |
| CV ± SD | 3.11 ± 0.66 | 1.63 ± 0.33 | 2.97 ± 0.62 | 3.40 ± 0.66 | 3.05 ± 0.65 |
| Gene (R) | ARF1 (0.98) | PTBP3 (0.97) | END04 (1.00) | 18S rRNA (1.00) | ARF2 (1.00) |
| CV ± SD | 3.17 ± 0.60 | 1.67 ± 0.32 | 3.21 ± 0.69 | 3.70 ± 0.78 | 3.39 ± 0.64 |
| Gene (R) | UFD1 (0.96) | 18S rRNA (0.99) | IF4E2 (1.00) | ARF2 (1.00) | PTBP3 (0.98) |
| CV ± SD | 3.35 ± 0.70 | 1.69 ± 0.35 | 3.25 ± 0.65 | 3.89 ± 0.76 | 3.47 ± 0.65 |
| Gene (R) | EF1α (0.95) | ARF1 (0.99) | RPAB5 (0.99) | UFD1 (1.00) | UFD1 (0.99) |
| CV ± SD | 3.36 ± 0.64 | 1.82 ± 0.34 | 3.66 ± 0.70 | 3.92 ± 0.83 | 3.53 ± 0.73 |
| Gene (R) | RPAB5 (0.98) | END04 (1.00) | UFD1 (0.67) | NUB1 (0.98) | ENDO4 (1.00) |
| CV ± SD | 3.49 ± 0.66 | 1.95 ± 0.41 | 3.70 ± 0.78 | 3.98 ± 0.84 | 3.54 ± 0.75 |
| Gene (R) | UBE4 (0.92) | IF4E2 (1.00) | UBE4 (0.99) | UBE4 (1.00) | 18S rRNA (0.99) |
| CV ± SD | 3.54 ± 0.71 | 2.08 ± 0.41 | 3.91 ± 0.80 | 4.05 ± 0.83 | 3.63 ± 0.74 |
| Gene (R) | IF4E2 (0.99) | NUB1 (0.95) | ARF2 (1.00) | ARF1 (1.00) | ERF3A (0.98) |
| CV ± SD | 3.55 ± 0.70 | 2.18 ± 0.44 | 3.96 ± 0.77 | 4.12 ± 0.79 | 3.73 ± 0.69 |
| Gene (R) | ARF2 (0.96) | UBE4 (0.99) | EF1α (0.99) | RPAB5 (0.98) | IF4E2 (0.99) |
| CV ± SD | 3.66 ± 0.70 | 2.24 ± 0.45 | 4.66 ± 0.89 | 4.22 ± 0.81 | 3.97 ± 0.77 |
| Gene (R) | NUB1 (0.98) | EF1α (0.94) | NUB1 (1.00) | IF4E2 (0.99) | RPAB5 (0.99) |
| CV ± SD | 3.85 ± 0.80 | 2.44 ± 0.46 | 4.69 ± 0.98 | 4.24 ± 0.85 | 4.09 ± 0.77 |
| Gene (R) | ERF3A (0.98) | ERF3A (0.99) | ERF3A (0.99) | ERF3A (0.98) | NUB1 (0.98) |
| CV ± SD | 3.98 ± 0.74 | 2.72 ± 0.50 | 4.74 ± 0.89 | 4.35 ± 0.82 | 4.18 ± 0.86 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, A.; Shao, D.; Cui, B.; Tong, X.; Zheng, Y.; Sun, J.; Li, H. Screening the Reference Genes for Quantitative Gene Expression by RT-qPCR During SE Initial Dedifferentiation in Four Gossypium hirsutum Cultivars that Have Different SE Capability. Genes 2019, 10, 497. https://doi.org/10.3390/genes10070497
Cao A, Shao D, Cui B, Tong X, Zheng Y, Sun J, Li H. Screening the Reference Genes for Quantitative Gene Expression by RT-qPCR During SE Initial Dedifferentiation in Four Gossypium hirsutum Cultivars that Have Different SE Capability. Genes. 2019; 10(7):497. https://doi.org/10.3390/genes10070497
Chicago/Turabian StyleCao, Aiping, Dongnan Shao, Baiming Cui, Xuecheng Tong, Yinying Zheng, Jie Sun, and Hongbin Li. 2019. "Screening the Reference Genes for Quantitative Gene Expression by RT-qPCR During SE Initial Dedifferentiation in Four Gossypium hirsutum Cultivars that Have Different SE Capability" Genes 10, no. 7: 497. https://doi.org/10.3390/genes10070497
APA StyleCao, A., Shao, D., Cui, B., Tong, X., Zheng, Y., Sun, J., & Li, H. (2019). Screening the Reference Genes for Quantitative Gene Expression by RT-qPCR During SE Initial Dedifferentiation in Four Gossypium hirsutum Cultivars that Have Different SE Capability. Genes, 10(7), 497. https://doi.org/10.3390/genes10070497





