Abstract
The incidence of thyroid cancer (TC), particularly well-differentiated forms (DTC), has been rising and remains the highest among endocrine malignancies. Although ionizing radiation (IR) is well established on DTC aetiology, other environmental and genetic factors may also be involved. DNA repair single nucleotide polymorphisms (SNPs) could be among the former, helping in explaining the high incidence. To further clarify the role of DNA repair SNPs in DTC susceptibility, we analyzed 36 SNPs in 27 DNA repair genes in a population of 106 DTCs and corresponding controls with the aim of interpreting joint data from previously studied isolated SNPs in DNA repair genes. Significant associations with DTC susceptibility were observed for XRCC3 rs861539, XPC rs2228001, CCNH rs2230641, MSH6 rs1042821 and ERCC5 rs2227869 and for a haplotype block on chromosome 5q. From 595 SNP-SNP combinations tested and 114 showing relevance, 15 significant SNP combinations (p < 0.01) were detected on paired SNP analysis, most of which involving CCNH rs2230641 and mismatch repair variants. Overall, a gene-dosage effect between the number of risk genotypes and DTC predisposition was observed. In spite of the volume of data presented, new studies are sought to provide an interpretability of the role of SNPs in DNA repair genes and their combinations in DTC susceptibility.
1. Introduction
Thyroid cancer (TC) is the most common endocrine malignancy and its increasing incidence raises concern. It is two to four times more frequent in women than in men and one of the most common malignancies in adolescent and young adults, ages 15–39 years, the median age at diagnosis being lower than that for most other types of cancer [1,2]. Papillary (PTC) and follicular (FTC) thyroid cancer, representing 85–90% and 5–10% of cases, respectively, are the most common histological varieties and are often collectively referred to as well-differentiated thyroid carcinoma (DTC). In contrast to anaplastic thyroid cancer (ATC), DTC prognosis is generally good, with high long-term survival and low disease-specific mortality [3,4].
DTC aetiology is multifactorial, resulting from the interplay between genetic and environmental factors: exposure to ionizing radiation (IR), particularly during childhood, remains the best-established modifiable risk factor, despite others – such as dietary habits (e.g., iodine intake), obesity and xenobiotic exposure – have also been proposed [2,4,5]. The importance of hereditary factors on DTC susceptibility is evidenced from familial studies demonstrating high disease risk among first-degree relatives and placing DTC as one of the cancers with higher heritability [6]. So far, the most robust evidence – provided by several genome wide association studies (GWASs), with independent replication across different populations – establishes markers at 9q22.33 (FOXE1), 14q13.3 (NKX2-1), 2q35 (DIRC3), 8p12 (NRG1) and 1q42.2 (PCNXL2) as the strongest genetic susceptibility markers for DTC (reviewed in [6,7]). Further candidate markers such as single nucleotide polymorphisms (SNPs) within genes involved in cell cycle control and apoptosis, DNA repair, intracellular signalling and transcriptional regulation have been proposed (reviewed in [8,9,10]) but many of these findings have not been properly replicated. Overall, currently proposed DTC risk markers are still largely insufficient to explain the high heritability of DTC [6]. It is possible that other, yet unidentified, genetic variants have a relevant impact on DTC susceptibility and thus explain part of the missing heritability of the disease. Their identification is therefore highly desirable.
DNA repair safeguards genomic integrity upon exposure to genotoxic agents, its absence or impairment leading to cancer-driving mutations in oncogenes or tumour suppressor genes (reviewed in [11,12]). A great number of DNA repair SNPs has been associated with cancer susceptibility (reviewed in [12,13]), strongly suggesting that such variants may, if functionally significant, modulate the individual sensitivity to genotoxic agents and, hence, contribute to cancer predisposition.
Considering the important role that IR and, possibly, other DNA damaging agents play in DTC aetiology, DNA repair SNPs could, through interference with DNA repair capacity, contribute to DTC susceptibility. Indeed, prior studies by our team do suggest that SNPs across different DNA repair pathways – e.g., RAD51 and XRCC3 (HR pathway), CCNH (NER pathway) and MSH6 (MMR pathway) – may be implicated in TC (or, more specifically, DTC) predisposition [14,15,16,17,18]. Such studies add on to prior and subsequent work by other teams [8,12,19,20,21,22,23,24,25] that propose additional markers and reinforce the notion that DNA repair SNPs may contribute to DTC risk. However, besides being scarce, these studies provide only limited information on the impact of the studied SNP in specific subpopulations, e.g., male versus female patients or early-onset versus late-onset DTC. Considering the specificities of DTC regarding gender distribution and median age at diagnosis [1,2] such detailed analysis could prove useful. Although gene-gene interactions could be of utmost importance in the real context, possibly decisive, they have only seldom evaluated and, when considered [19,20,22,24,26,27], analyses were usually limited to the combined effect of SNPs in the same gene or in genes of the same pathway. DNA repair proteins functionally interact with each other, both within the same DNA repair pathway and across different pathways, establishing ground for additive or even multiplicative effects of different SNPs (irrespective of their pathway) on DNA repair activity and, hence, cancer risk. This has been previously demonstrated for other types of cancer such as breast cancer [28,29,30] and, most likely, also applies to DTC. Such hypothesis has not, to the best of our knowledge, been investigated, justifying the usefulness of assessing the effect of combined genotypes on DTC risk.
In the present work we grouped and analysed all studies performed by our group on a Caucasian Portuguese population [14,15,16,17,18]. Since the actual biological situation reflects the concerted action of various alleles in the repair of DNA lesions that may be carcinogenic, all the data was re-analysed in order to identify intra and inter-pathway genotype combinations and thus further characterize the potential contribution of those DNA repair SNPs to DTC susceptibility. Such screening efforts may allow the identification of candidate SNPs for future use as susceptibility biomarkers, hence, the development of tailored DTC prevention policies and perhaps implementation of guidelines.
2. Material and Methods
2.1. Study Subjects
Overall, 335 Caucasian Portuguese subjects were enrolled in this hospital-based case-control study: 106 histologically confirmed DTC patients were recruited in the Service of Nuclear Medicine of the Portuguese Oncology Institute, Lisbon, Portugal where they were treated according to the hospital current practice and 229 unrelated age (±2 years) and gender-matched controls (two for each DTC case, in each of the previously published studies) were recruited at the Department of Clinical Pathology of the São Francisco Xavier Hospital, West Lisbon Hospital Centre, Portugal where they were seeking healthcare for non-neoplastic pathology. None of the study participants had personal history of prior malignancy nor familial history of thyroid disease.
In order to verify eligibility criteria and to account for potential confounding factors, information on demographic characteristics (e.g., gender, age, occupation), family history of cancer, lifestyle habits (e.g., smoking, alcohol drinking) and IR exposure was collected from each study participant, on recruitment, through a pre-designed questionnaire performed by trained interviewers. Prior exposure to relevant levels of ionizing radiation (i.e., other than that from natural and standard diagnostic sources) was denied by all subjects included in the study. Former smokers were considered as non-smokers if they gave up smoking 2 years before DTC diagnosis or 2 years before their inclusion as controls. The response rate was >95% for both cases and controls.
All studies were previously approved by the local ethics boards of the involved institutions and conducted in compliance with the Helsinki Declaration. On recruitment, prior to blood withdrawal, all eligible subjects were informed about the objectives of the study. Those agreeing to participate gave their written informed consent and were enrolled in the study. The anonymity of all participants was guaranteed.
2.2. SNP Selection
The selection of SNPs for genotyping was performed according to criteria that were predefined individually for each original study [14,15,16,17,18]. Briefly, eligible SNPs were required to exhibit a minor allele frequency (MAF) greater than 0.05 in Caucasian populations, the remaining criteria (e.g., being located in a coding or splice region, altering the amino acid sequence, being a tagging SNP, having been previously referred to in MEDLINE) varying according to the individual study, as indicated in the original studies of individual alleles.
Overall, a total of 36 DNA repair SNPs across all DNA repair pathways were selected for genotyping and analysed. Details on the genomic location, base and amino acid exchange and MAF of selected SNPs are presented on Table 1.

Table 1.
Selected SNPs and detailed information on the corresponding base and amino acid exchanges, minor allele frequency (MAF) and AB assay used for genotyping.
2.3. Practical Methodologies—Brief Description
All DNA samples were obtained after collection of peripheral venous blood samples from each participant. The DNA extraction was performed as described previously [14,15,16,17,18] using a commercial available kit (QIAamp® DNA mini kit; Qiagen GmbH, Hilden, Germany), according to the manufacturer’s recommendations. All samples were stored at −20 °C until further analysis.
Genotyping was carried out through either real-time polymerase chain reaction (PCR) or conventional PCR-restriction fragment length polymorphism (RFLP) techniques, as described in previous studies [14,15,16,17,18]. For real-time PCR—the option for the vast majority of SNPs considered in this study – genotyping was performed on an ABI 7300 Real-Time PCR system thermal cycler (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), using the commercially available TaqMan® SNP Genotyping Assays (Applied Biosystems) identified in Table 1. Conventional techniques of polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) were employed to genotype XRCC1 rs1799782, XRCC1 rs25487 and OGG1 rs1052133 (BER pathway); XPC rs2228000 and XPC rs2228001 (NER pathway); and XRCC3 rs861539 and XRCC2 rs3218536 (HR pathway). Primer design methods and sequences, PCR conditions, PCR product sizes, restriction analysis conditions and expected digestion pattern for each genotype have been described in full detail elsewhere [14,16,17] and will therefore not be reproduced here. Irrespective of the genotyping method, all inconclusive samples were reanalysed. Also, for quality control, at least 10–15% of genotype determinations were run in duplicates through independent experiments, with 100% concordance between experiments.
2.4. Statistical Analysis
Prior to analysis, genotype distributions for each studied SNP were checked for deviation from Hardy–Weinberg equilibrium (HWE) using SNPstat platform [31], in both case and control populations. Variable transformation was applied to categorize the only continuous variable (age of diagnosis) and the Chi-square test was then used to evaluate differences in genotype frequency, smoking status, age class and gender distributions between DTC patients and controls. Whenever the construction of 2 × 2 contingency tables was possible, the two-sided Fisher’s exact test was employed instead of the Chi-square test.
Logistic regression was used to estimate the risk of DTC associated with each genotype: risk estimates were calculated under the codominant, dominant and recessive models and expressed as crude and adjusted odds ratios (OR) and corresponding 95% confidence intervals (CI). Whenever adjustment was performed, terms for gender (male/female), age class (<30, 30–49, 50–69 and ≥70 years) and smoking habits (smokers/non-smokers) were included in the model, the most common homozygous genotype, female gender, lower age group and non-smoking status being considered the reference classes for such calculations. As data on prior IR exposure was not suitable for rigorous quantitative transformation, it was not possible to include such term in the adjustment model. Risk estimates were calculated in the whole population and after stratification according to histological type of tumour (papillary or follicular TC), gender (male and female) and age (<50 and ≥50 years).
Finally, the joint effect of multiple SNPs on DTC risk was estimated from application of logistic regression analysis (1) to relevant haplotypes, (2) to individual genetic risk scores calculated from genotype variables significant on single SNP analysis and (3) to all possible 2 × 2 combinations of the DNA repair SNPs included in this study. For the purpose of risk score calculations, genotypes presenting significant results on single SNP analysis were attributed a +1 score, the risk score for each participant corresponding to the sum of such scores. Samples with one or more missing genotypes were excluded from these calculations to avoid bias due to missing data. For paired SNP analysis, the combination of the most common homozygous genotypes of each individual SNP in the control group was taken as the reference category in OR calculations. Also, paired genotypes with frequency <5% in the study population were pooled together.
This is not a conclusive final study but an exploratory one that should be regarded as ‘proof of concept’. As such, the Bonferroni adjustment was deemed as not necessary as it is too conservative. Also, the complement of the false negative rate β to compute the power of a test (1−β) was not taken into account at this stage since further studies with more patients and controls should be undertaken to change over this preliminary study into a confirmatory positive one. All statistical analyses were performed with SPSS 22.0 (IBM SPSS Statistics for Windows, version 22.0, IBM Corp, Armonk, NY, USA) except for assessment of HWE deviation, MAF calculations, haplotype estimation and linkage disequilibrium (LD) analysis which were carried out using SNPstats [31]. Results were considered significant when the corresponding two-tailed p-values were <0.05 except for paired SNP analysis where, because of the high number of SNP-SNP combinations being tested, a more stringent significance level (p < 0.01) was employed. The study was approved by the Ethical Committee of Nova Medical School, Faculty of Medical Sciences with the number 05/2008 dated of January 9th, 2008. The approval was also obtained by the ethical committee of Portuguese Oncology Institute (IPO), the hospital responsible for blood samples collection with the reference GIC/357 dated of July 14th 2004.
3. Results
3.1. General Analysis
The general characteristics of the 106 DTC patients and their 229 age- and gender-matched controls included in this study are depicted in Table 2. The overall mean age of the study population was 51 years (52.1 in the patient group and 51.0 in the control group). As expected from the worldwide gender distribution for DTC [1,2], female patients greatly outnumbered male patients in the case group. Twelve (11.3%) DTC patients were categorized as smokers. Age distribution, gender and smoking habits were not significantly different between case and control populations. Concerning histological classification of tumours, 78 (73.6%) patients were diagnosed as papillary TC while 28 (26.4%) presented follicular tumours, in line with DTC histotype distributions commonly reported in the literature [4]. Three additional cases of poorly differentiated TC were also present in some of our original studies but, since this study concerns only with DTC, such cases (and the corresponding controls) were excluded from this analysis. Prior IR exposure (except for diagnostic X-rays) was denied by all cases.

Table 2.
General characteristics for the DTC case (n = 106) and control (n = 229) populations.
3.2. All DTC Cases
Allelic and genotypic frequencies as well as crude/adjusted ORs were calculated for all 36 DNA repair SNPs analysed in our study. Significant findings are reported in Table 3. The allelic and genotypic frequencies observed in the control group were in agreement with those expected for Caucasian populations. Also, for the majority of SNPs, genotype distributions were in Hardy-Weinberg equilibrium (HWE, p ≥ 0.05), in both case and control populations. Significant deviations from HWE were observed for OGG1 rs1052133, MUTYH rs3219489 and CDK7 rs2972388 in the control group and for XRCC1 rs1799782, XPC rs2228000 and MSH3 rs184967 in the DTC group. Further, strong linkage disequilibrium was observed between XRCC5 rs1051677 and rs6941, but not between any other pair of SNPs. XRCC5 rs6941 was thus excluded from further analysis, the conclusions taken for XRCC5 rs1051677 being valid for XRCC5 rs6941, since they behave as tag SNPs.

Table 3.
Genotype distribution in case and control populations and associated DTC risk (crude and adjusted ORs). Only SNPs presenting significant findings are shown.
As expected, both the comparison of genotype frequency distributions between case and control populations and the logistic regression analysis (Table 3) yielded results similar to those previously reported [14,15,16,17,18]: significant differences on the distribution of genotypic frequencies between cases and controls were observed for CCNH rs2230641 (p = 0.037 on the codominant model and p = 0.024 on the dominant model), for MSH6 rs1042821 (p = 0.042, on the codominant model and p = 0.037 on the recessive model) and for XRCC3 rs861539 (p = 0.021 on the codominant model and p = 0.011 on the recessive model). On logistic regression analysis, after adjustment for age, gender and smoking status, DTC risk was significantly increased in CCNH rs2230641 heterozygotes (adjusted OR = 1.89, 95% CI: 1.14–3.14, p = 0.014) and also in variant allele carriers, according the dominant model (adjusted OR = 1.79, 95% CI: 1.09–2.93, p = 0.021), in MSH6 rs1042821 variant allele homozygotes (adjusted OR = 3.42, 95% CI: 1.04-11.24, p = 0.042 on the codominant model; adjusted OR = 3.84, 95% CI: 1.18–12.44, p = 0.025 on the recessive model), in XRCC3 rs861539 variant allele homozygotes (adjusted OR = 2.20, 95% CI: 1.20–4.03, p = 0.011 on the recessive model) and in XPC rs2228001 variant allele homozygotes (adjusted OR = 1.97, 95% CI: 1.01–3.84, p = 0.046 on the recessive model). A borderline significant DTC risk reduction was observed in ERCC5 rs2227869 heterozygotes (adjusted OR = 0.39, 95% CI: 0.16-1.00, p = 0.049). The association between XPC rs2228001 and DTC risk is a new finding emerging from this reanalysis, since the recessive model of inheritance had not been applied in the original study [17]. No additional significant differences in genotype frequency distributions nor associations with DTC risk were found, irrespective of the model assumed.
3.3. Stratified Analysis
Stratified analysis according to histological tumour type, gender and age may be important to identify any subgroup-specific risk association but was only partially performed in prior studies in this population. On stratification according to histological criteria (Table 4), this study confirmed prior observations [14,17,18] of increased papillary TC risk in XPC rs2228001 and XRCC3 rs861539 variant allele homozygotes (XPC rs2228001: adjusted OR = 2.31, 95% CI: 1.07–4.98, p = 0.033; XRCC3 rs861539: adjusted OR = 2.10; 95% CI: 1.07–4.11; p = 0.031, both on the recessive model), decreased papillary TC risk in ERCC5 rs2227869 heterozygotes (adjusted OR = 0.23, 95% CI: 0.07–0.81, p = 0.022, on the codominant model) or variant allele carriers (adjusted OR = 0.22, 95% CI: 0.06–0.77, p = 0.018, on the dominant model) and increased follicular TC risk in MLH3 rs175080 variant allele carriers (crude OR = 3.95, 95% CI: 1.05–14.81, p = 0.042) and MSH6 rs1042821 variant allele homozygotes (adjusted OR = 20.98, 95% CI: 1.08-406.53, p = 0.044, on the codominant model; adjusted OR = 23.70, 95% CI: 1.25–449.32, p = 0.035, on the recessive model). Interestingly, three other significant associations were observed in this reanalysis that were not present or had not been detected in the original studies, while two previously observed associations were lost in this reanalysis: a previously undetected decreased papillary TC risk was observed in MUTYH rs3219489 heterozygotes (crude OR = 0.56, 95% CI: 0.32–1.00, p = 0.048) and variant allele carriers (crude OR = 0.57, 95% CI: 0.33–0.99, p = 0.048) as well as in NBN rs1805794 variant allele homozygotes (adjusted OR = 0.28, 95% CI: 0.08-0.97, p = 0.045, on the recessive model) while the presence of the variant allele of XRCC2 rs3218536 exhibited a protective effect for follicular TC (crude OR = 0.21, 95% CI: 0.04–1.00, p = 0.049, either for heterozygotes in the codominant model and for variant allele carriers in the dominant model). In contrast, the associations of XRCC5 rs2440 and CCNH rs2230641 genotypes with papillary and follicular TC risk, respectively, reported in our original studies [15,17], were no longer observed.

Table 4.
Genotype distribution in the case population (n = 106) and associated DTC risk (crude and adjusted ORs), after stratification according to histological type, gender and age. Only SNPs presenting significant findings are shown.
On gender stratification (Table 4), when considering female patients only, a significantly increased DTC risk was evident for CCNH rs2230641 heterozygotes (adjusted OR = 1.97, 95% CI: 1.13–3.43, p = 0.017) and variant allele carriers (adjusted OR = 1.90, 95% CI: 1.11–3.24, p = 0.020), for XPC rs2228001 variant allele homozygotes (adjusted OR = 2.00, 95% CI: 1.01–3.96, p = 0.048, on the recessive model), for MSH6 rs1042821 variant allele homozygotes (adjusted OR = 4.78, 95% CI: 1.17–19.56, p = 0.030, on the codominant model; adjusted OR = 5.42, 95% CI: 1.34–21.92, p = 0.018, on the recessive model) and for XRCC3 rs861539 variant allele homozygotes (adjusted OR = 2.36, 95% CI: 1.12–4.97, p = 0.024, on the codominant model; adjusted OR = 2.68, 95% CI: 1.39–5.18, p = 0.003, on the recessive model). Opposing, ERCC5 rs2227869 heterozygotes (adjusted OR = 0.25, 95% CI: 0.07–0.88, p = 0.030) and variant allele carriers (adjusted OR = 0.32, 95% CI: 0.11–0.97, p = 0.044) as well as ERCC5 rs17655 variant allele homozygotes (adjusted OR = 0.27, 95% CI: 0.08–0.95, p = 0.041, on the recessive model) presented a significant risk reduction among female patients. Among these gender-specific genetic effects, only the association with MSH6 rs1042821 had been reported in the original studies [18]. No significant association was observed in the male subset of patients, possibly because of the low number of cases in this gender group. An association between XRCC5 rs1051677 and TC risk had previously been identified in this subset of patients [15] but significance was lost upon restricting analysis to well-differentiated forms of TC (this study).
Stratified analysis according to the age of diagnosis had only been performed in some of our initial studies, namely those involving SNPs of the BER and MMR pathways [16,18], with negative results. We therefore extended this analysis to the remaining DNA repair SNPs, considering two age groups: <50 and ≥50 years. In patients under 50 years of age, both homozygosity for the XPC rs2228001 variant allele (adjusted OR = 2.86, 95% CI: 1.01–8.08, p = 0.048, on the recessive model) and the presence of at least one XRCC5 rs2440 variant allele (adjusted OR = 2.53, 95% CI: 1.02–6.26, p = 0.045) were associated with increased DTC risk. When restricting the analysis to patients with 50 or more years of age, DTC risk was increased in CCNH rs2230641 heterozygotes (adjusted OR = 2.91, 95% CI: 1.51–5.60, p = 0.001) and variant allele carriers (adjusted OR = 3.04, 95% CI: 1.59–5.81, p = 0.001), in RAD51 rs1801321 variant allele homozygotes (adjusted OR = 2.99, 95% CI: 1.25-7.14, p = 0.014, on the codominant model; unadjusted OR = 2.03, 95% CI: 1.00–4.12, p = 0.049, on the recessive model) and variant allele carriers (adjusted OR = 2.14, 95% CI: 1.06–4.32, p = 0.034) and in XRCC3 rs861539 variant allele homozygotes (adjusted OR = 2.63, 95% CI: 1.16–5.97, p = 0.021, on the recessive model). On the contrary, the presence of at least one variant ERCC6 rs2228529 allele (adjusted OR = 0.47, 95% CI: 0.24–0.92, p = 0.028) and its presence in heterozygosity (adjusted OR = 0.48, 95% CI: 0.24–0.97, p = 0.042) were associated with a DTC risk reduction in this older age group.
No further correlations between individual DNA repair SNPs and DTC risk were observed on histology-, gender- and age-based stratification analysis.
3.4. Combined Genotypes
In order to investigate the joint effect of multiple SNPs on DTC risk, genetic risk scores (RS) were calculated for each study participant, considering only significant findings on single SNP analysis. As depicted in Table 5, after adjusting for covariates, DTC risk was more than two and five times higher in individuals bearing, respectively, 2 (adjusted OR = 2.68, 95% CI: 1.56–4.59, p < 0.001) and 3 or more (adjusted OR = 5.02, 95% CI: 2.24–11.24, p = 0.001) risk genotypes (CCNH rs2230641 Val/Ala or Ala/Ala; ERCC5 rs2227869 Cys/Cys or Ser/Ser; XPC rs2228001 Gln/Gln; MSH6 rs1042821 Glu/Glu; XRCC3 rs861539 Met/Met), when compared to individuals bearing none or only one of such risk genotypes. Similar associations between RS and TC risk were also observed on stratification according to histological, gender or age criteria, after adapting RS calculations to the SNPs significant for each strata (Table 5). A high significance level was observed in most cases (p < 0.001 in approximately 50% of RS categories) and was even greater if higher RS categories were merged together (results not shown).

Table 5.
Risk score (RS) in case and control populations and associated DTC risk (crude and adjusted ORs). Risk scores calculated from significant results on single SNP analysisa.
Also, in order to investigate the combined effect of different pairs of SNPs on DTC risk, we performed a paired SNP analysis considering all possible 2 × 2 combinations of the DNA repair SNPs included in this study. Overall, 595 SNP-SNP combinations were tested, 114 (approximately 20%) of which yielded significant results at a 0.05 significance level (results not shown). Considering that such a high number of hypothesis being tested may result in a considerable number of false positive findings, a more stringent significance level (p < 0.01) was employed in this analysis, limiting the number of SNP pairs with significant findings to 15 (approximately 2.5% of all possible combinations). Such significant findings are depicted in Table 6 and also in Figure 1. CCNH rs2230641 emerges from Figure 1 as the DNA repair SNP most frequently represented in significant SNP-SNP combinations, both at 0.01 and 0.05 significance levels, followed by RAD51 rs1801321, MLH3 rs175080 and MSH4 rs5745549 (0.01 significance level) or RAD51 rs1801321and XRCC3 rs861539 (0.05 significance level). MMR variants were the most frequently involved as they were present in 9 of the 15 SNP-SNP combinations that were significant. Also, among significant findings, 3 intra-pathway SNP combinations were detected: RAD51 rs1801321–XRCC3 rs861539 (HR pathway), MLH3 rs175080–MSH6 rs1042821 and MSH4 rs5745549–MSH6 rs1042821 (MMR pathway).

Table 6.
Two-way SNP interactions among DNA repair genes: distribution of combined genotypes in enrolled populations and associated DTC risk (adjusted ORs). Only SNPs presenting significant findings (p < 0.01) are shown.
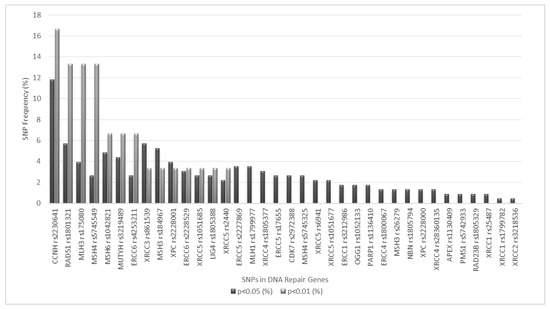
Figure 1.
SNP frequency (%) in SNP-SNP pairs showing significant results at p < 0.01 and p < 0.05 levels. Only SNPs presenting significant results (p < 0.05) on combined genotype analysis are shown.
Finally, haplotype analysis was applied to SNPs located in the same chromosome arm, since these are likely to segregate together. According to such criteria, it was possible to establish 8 blocks of DNA repair SNPs, of which only one, located on chromosome 5q and comprising 6 SNPs (CCNH rs2230641, CDK7 rs2972388, MSH3 rs26279, MSH3 rs184967, XRCC4 rs1805377 and XRCC4 rs28360135), revealed significant associations with DTC (Table 7): two different allele combinations were associated with a significantly decreased DTC risk, when compared to the most frequent combination of chromosome 5q SNPs (adjusted OR1 = 0.26, 95% CI: 0.08–0.87, p = 0.030; adjusted OR2 = 0.15, 95% CI: 0.03–0.72, p = 0.019). Haplogroup analysis comprising all SNPs under study could also prove useful to understand the joint effect of the variants since it would better reflect the real context situation (where different DNA repair proteins interact with each other) but could not be performed because, considering the high number of SNPs under study, the frequency of each specific allele combination would be too low for meaningful results to be obtained.

Table 7.
Haplotypes comprising SNPs located in the same chromosome arm and corresponding DTC risk (adjusted ORs). Only haplotypes presenting significant results are shown.
4. Discussion
In order to further characterize the potential contribution of DNA repair SNPs to DTC susceptibility, we aggregated and reanalysed the data from our previously published case-control studies [14,15,16,17,18] performed on a Caucasian Portuguese population.
A significant risk increase was observed, after adjustment for age, gender and smoking status, in CCNH rs2230641 heterozygotes and variant allele carriers, in MSH6 rs1042821 variant allele homozygotes (codominant and recessive model), in XRCC3 rs861539 variant allele homozygotes (recessive model) and in XPC rs2228001 variant allele homozygotes (recessive model), while the heterozygous ERCC5 rs2227869 genotype was associated with a borderline risk reduction. Except for XPC rs2228001, which is a new finding emerging from this reanalysis because the recessive model of inheritance had not been applied in the original study, such results are fundamentally similar to those reported on the original studies despite, on reanalysis, data was restricted to DTC cases and corresponding controls. A role for these variants specifically on well-differentiated forms of TC is thus apparent from this reanalysis. As these findings have been discussed in detail in the original studies, they will be discussed here only briefly, with emphasis on new data published since then.
XRCC3 participates in HR to maintain chromosome stability and repair DNA damage and is therefore a highly suspected candidate gene for cancer susceptibility. The XRCC3 rs861539 has been the most studied genetic variant of XRCC3 gene, especially because is located in a functional relevant domain of the protein, in an interaction region with other proteins such as RAD51 [22,32]. The presence of this variant may affect the structure of this DNA repair protein and lead to a deficiency in the HR pathway. As a result, the HR pathway may be compromised, shifting the repair mechanism to NHEJ, promoting chromosome instability and disturbing the cellular repair capacity [33]. The potential contribution of XRCC3 rs861539 to cancer susceptibility has been widely addressed: while conflicting evidence exists, several large meta-analyses strongly support a positive association with cancer susceptibility, namely breast [34,35,36] and bladder cancer [36,37,38], among others. In the particular context of thyroid cancer, interestingly, multiple studies [22,39,40,41,42,43], including a meta-analysis [44], have suggested the XRCC3 rs861539 variant T allele and/or, in particular, the TT homozygous genotype to be associated with increased risk of TC or, more specifically, PTC. In another meta-analysis [45] such association was also detected but only in Caucasian populations. Therefore, despite studies reporting no significant association also exist [46,47], the vast majority of available evidence supports our results and suggests a role for XRCC3 rs861539 in DTC susceptibility.
To the best of our knowledge, none of the remaining SNPs presenting significant results on overall analysis has been evaluated in the context of DTC (or TC) susceptibility.
XPC codes for a DNA binding protein that acts forming the distortion-sensing component of NER by binding tightly with another important NER protein, HR23B, to form a stable XPC-HR23B complex, thus playing a central role in the process of early damage recognition [48,49]. XPC-HR23B complex can recognize a variety of DNA adducts formed by exogenous carcinogens and binds to the DNA damage sites. Therefore, it may play a role in decreasing the toxic effects of such carcinogens and its deficiency may interact with carcinogen exposure [50]. XPC is also involved in DNA damage-induced cell cycle checkpoint regulation and apoptosis, removal of oxidative DNA damage and redox homeostasis [49,51]. XPC rs2228001 (an A-to-C transition in exon 15) leads to a substitution of glutamine for lysine in codon 939 (Lys939Gln) and is located in the domain interacting with the transcription factor IIH (TFIIH) complex [50,52,53,54,55], initiating the global genome NER pathway. XPC rs2228001 is one of the most extensively studied NER pathway SNPs, as numerous case-control association studies and meta-analyses have been performed to investigate its potential role on cancer predisposition. In line with our data for DTC, a modest but consistent association of the Gln/Gln homozygous genotype with overall cancer risk is apparent from two of the three meta-analysis that pool data from different cancer types [56,57,58]. Evidence from these and other cancer site-specific meta-analyses is stronger for lung [53,56,57,58,59,60], bladder [54,56,61,62] and colorectal cancer (CRC) [56,58] [63,64], but also exists for other cancer types such as upper digestive system cancer [65] and hepatocellular carcinoma [50,66]. XPC rs2228001 genotype has also been found to correlate with survival of hepatocellular patients [66], with XPC mRNA expression levels [60,66,67], with drug-induced toxicity in cancer patients treated with platinum-based chemotherapeutic agents (e.g., cisplatin) [68,69], with sensitivity of lung squamous cell carcinoma patients to chemotherapy [67] and to interfere with the capacity to repair DNA lesions induced by, e.g., benzo(a)pyrene [70,71,72], gamma-radiation [70], X-rays [73], UV radiation [74], aflatoxin B1 [50] and meat-derived carcinogens [75]. Overall, evidence strongly suggests that XPC rs2228001 genotype is associated with altered DNA repair capacity, establishing ground for a putative role of this SNP in cancer susceptibility.
The MSH6 gene (mutS homolog 6) is a member of a set of genes known as the mismatch repair (MMR) genes. MSH6 integrates the MutSα complex, a sensor of genetic damage that, besides its role in the repair of replication errors, cooperates with other DNA repair and damage-response signalling pathways to allow for cell cycle arrest, DNA repair and/or apoptosis of genetically damaged cells. Several MSH6 mutations have been identified and suggested as causative in Lynch syndrome (LS) patients [76,77,78,79,80]. Despite TC is not part of the usual LS spectrum, the effect of MSH6 in TC susceptibility has previously been explored [81,82]. MSH6 rs1042821 has also been frequently investigated in the context of cancer susceptibility, mostly with inconclusive findings [83,84,85,86,87,88,89,90]. Consistent with our results, MSH6 rs1042821 has previously been associated with increased CRC risk [91,92,93], highly malignant bladder cancer [94], pancreatic cancer [95] and triple negative breast cancer (TNBC) [96]. On the contrary, the T allele [97] and the CT heterozygous genotype [98] have been associated with decreased colorectal and hepatocellular carcinoma, respectively. The only meta-analysis concerning the role of MSH6 rs1042821 on cancer predisposition that we are aware of is also inconclusive [99]. Despite plausible, a potential role for MSH6 rs1042821 on cancer predisposition (DTC, in particular) remains elusive. Further well-powered studies are needed to clarify this issue.
The role of CCNH rs2230641 on cancer predisposition has only seldom been evaluated: in agreement with our results, a significantly increased bladder cancer risk in ever smokers has been reported for C allele carriers [100] but, on the contrary, such genotype has also been associated with a significantly decreased risk of chronic leukaemia [101]. Most other studies, namely in oesophageal [102], bladder [103], biliary tract [104] and renal cell carcinoma [105], as well as in oral premalignant lesions [106] have been inconclusive. Interestingly, the pharmacogenomic implications of CCNH rs2230641 on the outcome of platinum-based chemotherapy have also been evaluated, results supporting a role for CCNH rs2230641 on the response to DNA damaging agents: the presence of the CCNH rs2230641 variant C allele has been associated with longer survival in NLCSC patients receiving platinum-based chemotherapy [107] and with increased incidence and severity of oxaliplatin-induced acute peripheral neuropathy in digestive tract cancer patients undergoing with the oxaliplatin-based chemotherapy [108]. Similarly, increased risk of severe oxaliplatin-induced acute peripheral neuropathy was observed by Custodio et al. [109] in high-risk stage II and stage III colon cancer patients homozygous for the C allele, submitted to oxaliplatin-based adjuvant chemotherapy. CCNH codes for a highly conserved cyclin protein that participates in several cellular processes such as the NER pathway, cell cycle regulation and receptor phosphorylation, among others [48,110]. Although data on the functional relevance of rs2230641 is lacking, the pleiotropic effects of CCNH confer biological plausibility to our hypothesis that CCNH variants may be involved in cancer susceptibility.
Finally, ERCC5, also known as XPG, is located on chromosome 13q22–q33 [111] and comprises 15 exons [112,113]. It encodes a structure-specific endonuclease that has multiple functions during NER [114], reason why defects in this gene can impair DNA repair resulting in genomic instability and carcinogenesis [115]. In fact, only a few studies have considered the putative contribution of ERCC5 rs2227869 to cancer susceptibility, most being inconclusive. Interestingly, the only significant findings reported thus far are in line with those reported here, suggesting a protective role for the heterozygous genotype: Hussain et al. [116] reported a significant reduction in stomach cancer risk in heterozygous genotype individuals and a similar, despite nonsignificant, trend has also been independently observed for melanoma [117] and for squamous cell carcinoma of the head and neck (SCCHN) [118]. More importantly, in the only meta-analysis performed to date [119], a decrease in cancer risk in ERCC5 rs2227869 heterozygotes (and for the C allele) has also been reported.
Many of these (and other) SNPs also presented significant findings on stratifying data according to hystotype, gender and age: on histological stratification, significant associations were observed between XRCC3 rs861539, XPC rs2228001, ERCC5 rs2227869, MUTYH rs3219489 and NBN rs1805794 and papillary TC, while MSH6 rs1042821, MLH3 rs175080 and XRCC2 rs3218536 were associated with follicular TC. XRCC3 rs861539, XPC rs2228001, MSH6 rs1042821, CCNH rs2230641, ERCC5 rs2227869 and ERCC5 rs17655 were associated with DTC in the female subset while no association was observed in males. Finally, XPC rs2228001 and XRCC5 rs2440 were associated with DTC in participants younger than 50 years, while, in participants aged 50 or more years, the DTC-associated SNPs included XRCC3 rs861539, CCNH rs2230641, ERCC6 rs2228529 and RAD51 rs1801321.
It is unclear whether these findings (and which among these) truly represent group-specific effects or whether they simply reflect the overall effect on the largest groups (i.e., when group sizes are unbalanced, e.g., papillary TC vs follicular TC, female vs male) and the corresponding lack of power to detect an effect on the smallest groups. Also, due to the low sample size on each strata, some of these results may simply represent incident findings (type I errors). XRCC3 rs861539, for example, has been previously associated with papillary TC [22,39,40]—in line with our results—but not with follicular TC. An effect of XRCC3 rs861539 genotype in follicular TC cannot, however, be excluded since follicular TC is much less frequent than papillary TC and these studies may have been underpowered to detect such effect. Also, Su et al. [120] have demonstrated the homozygous genotype of this SNP to be associated with breast cancer, the association being stronger in women younger than 55 years, with earlier first menarche or with latter menopause. This suggests an oestrogen-potentiated genetic effect, compatible with our own observation of increased DTC risk in XRCC3 rs861539 TT homozygotes among females but not among males. Further, the involvement of CCNH, through a cyclin-activated kinase complex, in oestrogen receptor phosphorylation [48] provides a possible rationale for our own observation of an association of the CCNH rs2230641genotype with DTC among females but not among males. Finally, the association of MSH6 rs1042821with DTC, observed in this study for female but not male individuals, is compatible with the growing evidence placing DTC as an oestrogen-associated cancer [121,122,123,124] and implicating MSH6 in such cancers [78,125,126,127,128,129]. These selected examples highlight the plausibility of the existence of group-specific genetic effects. Overall, such hystotype, gender and age specifies in DTC susceptibility are likely since (1) papillary and follicular TC represent distinct entities, with hystotype-specific molecular profiles (e.g., BRAF mutations and RET/PTC rearrangements in PTC, RAS mutations and PAX8/PPARγ translocations in FTC) [130]; (2) important gender differences exist in the incidence of DTC (i.e., DTC is, as previously stated two to four times more frequent in women than in men) [1,2]; and (3) DTC presents some age specificities, uncommon in other types of cancer (DTC is one of the most common malignancies in adolescent and young adults, the median age at diagnosis being lower than that for most other types of cancer) [1,2]. Further well-powered studies are urgently needed to clarify these results and thus establish which of these SNPs, if any, represents true group-specific susceptibility biomarkers.
Considering the multifactorial nature of DTC aetiology and the probable involvement of multiple genetic factors, alone or in combination, in DTC susceptibility, we undertook a combined genotype analyses to investigate the joint effect of multiple SNPs on DTC risk. When combining all risk genotypes significant at single SNP analysis into a unique unbalanced risk score, a clear-cut gene-dosage effect between the number of risk genotypes (unbalanced risk score) and DTC risk was observed, both on global analysis (considering all DTC cases and corresponding controls) and after stratification according to histological, gender and age criteria. This is biologically plausible since the different DNA repair proteins physically and functionally interact with each other, within the same or different DNA repair pathways, establishing ground for additive or even multiplicative effects of different SNPs on DNA repair activity and, hence, cancer risk. Such polygenic approach to assess the cumulative effects of multiple genetic variants on cancer risk has previously been employed [27,107,131,132], supporting its usefulness and clinical potential.
To investigate the effect of specific DNA repair SNP combinations on DTC risk, all possible 2 × 2 combinations were tested on paired SNP analysis, yielding fifteen SNP pairs with p < 0.01. Multiple interactions between SNPs from different DNA repair pathways and, even, other DNA damage response proteins have previously been reported [39,42,66,87], providing a rationale for such approach. Of notice, CCNH rs2230641 was the most frequently represented DNA repair SNP in such significant combinations, both at 0.01 and 0.05 significance levels, a finding that is compatible with the pleiotropic role of CCNH in DNA damage repair, cell cycle regulation and receptor phosphorylation [48,110]. More importantly, the contribution of MMR variants to the joint effect of DNA repair SNPs on DTC risk is evident from our results, as they were present in 9 of the 15 SNP pairs presenting significant findings. Besides its critical role in post-replication repair (through recognition and repair of base-base mispairs and insertion/deletion loops that arise during replication), the MMR pathway cooperates with other repair pathways in the recognition and subsequent repair of DNA damage induced by IR, UV light, oxidative stress or genotoxic chemicals (e.g., oxidative lesions, double strand breaks, pyrimidine dimers and inter-strand crosslinks) and contributes to damage-induced cytotoxicity through downstream signalling for cell cycle arrest and apoptosis [133,134,135]. Therefore, considering the large spectre of action of the MMR pathway, an elevated number of interactions between MMR and other DNA repair SNPs is expected. Such hypothesis, in line with our findings, has been recently strengthened by a report [136] associating SNPs from different DNA repair pathways with CRC in Lynch syndrome patients, a cancer predisposition condition originated by germline MMR mutations. Finally, among SNP pairs presenting significant findings in this study, three are intra-pathway combinations involving either HR or MMR pathway SNPs. The joint effects of MLH3 rs175080 – MSH6 rs1042821 and MSH4 rs5745549 – MSH6 rs1042821 (MMR pathway) SNP combinations were reported and discussed in our original study [18]. The joint effect of RAD51 rs1801321 and XRCC3 rs861539 (HR pathway) on cancer risk has been previously reported for breast cancer [137], in line with our results, and may be of particular relevance for DTC since the formation of radiation damage-induced RAD51 foci requires functional XRCC3 [138].
Finally, on applying haplotype analysis to SNPs that are located in the same chromosome arm (thus likely to segregate together), one block of DNA repair SNPs located on chromosome 5q (comprising CCNH rs2230641, CDK7 rs2972388, MSH3 rs26279, MSH3 rs184967, XRCC4 rs1805377 and XRCC4 rs28360135) was associated with DTC risk in our study. Such results further suggest an independent or interactive effect of these SNPs on DTC predisposition.
Overall, our results suggest that DNA repair SNPs across different pathways and may contribute to DTC predisposition, possibly exerting cumulative effects. This is of relevance since the estimated high heritability of DTC is only partially explained, even when considering the contribution of several GWAS recently performed. Gene-gene and gene-environment interactions have been hypothesised to play an important role so their identification and in-depth study is highly desirable to explain the “missing” heritability of DTC. However, the results presented here should be regarded only as proof of concept and must therefore be validated through replication in larger independent populations. Future studies should also be designed with the intention of accounting for environmental factors such as IR exposure and iodine deficiency (and their potential interaction with genetic factors). In addition, they should be sufficiently powered to allow other, less frequent but potentially relevant SNPs, to be studied and to allow more sophisticated and conclusive gene-gene interaction analysis to be performed. Finally, in order to strengthen our preliminary findings, the functional significance of these SNPs should be further investigated as well as their potential association with mutational events involved in DTC carcinogenesis (e.g., BRAF mutations and RET/PTC rearrangements).
Author Contributions
Conceptualization was mainly developed by J.R., T.C.F and E.L; Methodology was performed by, O.M.G. and J.R.; Validation proceedings by L.S.S., B.C.G. and S.N.S.; Formal Analysis was done by L.S.S. and S.N.S; Investigation was mainly performed by L.S.S., B.C.G. and H.N.B.; Resources acquired in restrict collaboration by O.M.G., T.C.F. and A.P.A..; Data Curation, O.M.G., T.C.F. and E.L.; Writing—Original Draft Preparation, L.S.S; Writing – Review & Editing, B.C.G., H.N.B., O.M.G., A.P.A., S.N.S., J.R.; Visualization has been prepared by L.S.S., and S.N.S.; Supervision of this project, J.R.; Project Administration, J.R. and E.L.; Funding Acquisition, J.R.
Funding
This work was supported by FCT – Fundação para a Ciência e a Tecnologia (Portuguese Foundation for Science and Technology) through Project UID/BIM/00009/2019-Centre for Toxicogenomics and Human Health.
Acknowledgments
The authors warmly acknowledge the generous collaboration of patients and controls in this study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer, 2018. Available online: https://gco.iarc.fr/today (accessed on 28 May 2019).
- Kitahara, C.M.; Sosa, J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016, 12, 646. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.V.; Osamura, R.Y.; Klӧppel, G.N.; Rosai, J. WHO Classification of Tumours of Endocrine Organs, 4th ed.; International Agency for Research on Cancer (IARC) Press: Lyon, France, 2017; Volume 10, pp. 65–144. [Google Scholar]
- Khosravi, M.H.; Kouhi, A.; Saeedi, M.; Bagherihagh, A.; Amirzade-Iranaq, M.H. Thyroid Cancers: Considerations, Classifications, and Managements. In Diagnosis and Management of Head and Neck Cancer; Akarslan, Z., Ed.; IntechOpen: London, UK, 2017; pp. 57–82. [Google Scholar] [CrossRef]
- Marcello, M.A.; Malandrino, P.; Almeida, J.F.M.; Martins, M.B.; Cunha, L.L.; Bufalo, N.E.; Pellegriti, G.; Ward, L.S. The influence of the environment on the development of thyroid tumors: A new appraisal. Endocr. Relat. Cancer 2014, 21, T235–T254. [Google Scholar] [CrossRef] [PubMed]
- Saenko, V.A.; Rogounovitch, T.I. Genetic Polymorphism Predisposing to Differentiated Thyroid Cancer: A Review of Major Findings of the Genome-Wide Association Studies. Endocrinol. Metab. 2018, 33, 164–174. [Google Scholar] [CrossRef]
- Hwangbo, Y.; Park, Y.J. Genome-Wide Association Studies of Autoimmune Thyroid Diseases, Thyroid Function, and Thyroid Cancer. Endocrinol. Metab. 2018, 33, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Adjadj, E.; Schlumberger, M.; de Vathaire, F. Germ-line DNA polymorphisms and susceptibility to differentiated thyroid cancer. Lancet. Oncol. 2009, 10, 181–190. [Google Scholar] [CrossRef]
- Iñigo, L.; Mercedes, R. Association studies in thyroid cancer susceptibility: Are we on the right track? J. Mol. Endocrinol. 2011, 47, R43–R58. [Google Scholar] [CrossRef]
- Figlioli, G.; Elisei, R.; Romei, C.; Melaiu, O.; Cipollini, M.; Bambi, F.; Chen, B.; Köhler, A.; Cristaudo, A.; Hemminki, K.; et al. A Comprehensive Meta-analysis of Case–Control Association Studies to Evaluate Polymorphisms Associated with the Risk of Differentiated Thyroid Carcinoma. Cancer Epidemiol. Biomark. Prev. 2016, 25, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Gatzidou, E.; Michailidi, C.; Tseleni-Balafouta, S.; Theocharis, S. An epitome of DNA repair related genes and mechanisms in thyroid carcinoma. Cancer Lett. 2010, 290, 139–147. [Google Scholar] [CrossRef]
- Köberle, B.; Koch, B.; Fischer, B.M.; Hartwig, A. Single nucleotide polymorphisms in DNA repair genes and putative cancer risk. Arch. Toxicol. 2016, 90, 2369–2388. [Google Scholar] [CrossRef]
- Bastos, H.N.; Antão, M.R.; Silva, S.N.; Azevedo, A.P.; Manita, I.; Teixeira, V.; Pina, J.E.; Gil, O.M.; Ferreira, T.C.; Limbert, E.; et al. Association of Polymorphisms in Genes of the Homologous Recombination DNA Repair Pathway and Thyroid Cancer Risk. Thyroid 2009, 19, 1067–1075. [Google Scholar] [CrossRef]
- Gomes, B.C.; Silva, S.N.; Azevedo, A.P.; Manita, I.; Gil, O.M.; Ferreira, T.C.; Limbert, E.; Rueff, J.; Gaspar, J.F. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol. Rep. 2010, 24, 1079–1085. [Google Scholar]
- Santos, L.S.; Branco, S.C.; Silva, S.N.; Azevedo, A.P.; Gil, O.M.; Manita, I.; Ferreira, T.C.; Limbert, E.; Rueff, J.; Gaspar, J.F. Polymorphisms in base excision repair genes and thyroid cancer risk. Oncol. Rep. 2012, 28, 1859–1868. [Google Scholar] [CrossRef]
- Santos, L.S.; Gomes, B.C.; Gouveia, R.; Silva, S.N.; Azevedo, A.P.; Camacho, V.; Manita, I.; Gil, O.M.; Ferreira, T.C.; Limbert, E.; et al. The role of CCNH Val270Ala (rs2230641) and other nucleotide excision repair polymorphisms in individual susceptibility to well-differentiated thyroid cancer. Oncol. Rep. 2013, 30, 2458–2466. [Google Scholar] [CrossRef]
- Santos, L.S.; Silva, S.N.; Gil, O.M.; Ferreira, T.C.; Limbert, E.; Rueff, J. Mismatch repair single nucleotide polymorphisms and thyroid cancer susceptibility. Oncol. Lett. 2018, 15, 6715–6726. [Google Scholar] [CrossRef]
- Bashir, K.; Sarwar, R.; Fatima, S.; Saeed, S.; Mahjabeen, I.; Akhtar Kayani, M. Haplotype analysis of XRCC1 gene polymorphisms and the risk of thyroid carcinoma. J. BUON 2018, 23, 234–243. [Google Scholar]
- Bashir, K.; Sarwar, R.; Saeed, S.; Mahjabeen, I.; Kayani, M.A. Interaction among susceptibility genotypes of PARP1 SNPs in thyroid carcinoma. PLoS ONE 2018, 13, e0199007. [Google Scholar] [CrossRef]
- Sandler, J.E.; Huang, H.; Zhao, N.; Wu, W.; Liu, F.; Ma, S.; Udelsman, R.; Zhang, Y. Germline Variants in DNA Repair Genes, Diagnostic Radiation, and Risk of Thyroid Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 285–294. [Google Scholar] [CrossRef]
- Sarwar, R.; Mahjabeen, I.; Bashir, K.; Saeed, S.; Kayani, M.A. Haplotype Based Analysis of XRCC3Gene Polymorphisms in Thyroid Cancer. Cell Physiol. Biochem. 2017, 42, 22–33. [Google Scholar] [CrossRef]
- Lonjou, C.; Damiola, F.; Moissonnier, M.; Durand, G.; Malakhova, I.; Masyakin, V.; Le Calvez-Kelm, F.; Cardis, E.; Byrnes, G.; Kesminiene, A.; et al. Investigation of DNA repair-related SNPs underlying susceptibility to papillary thyroid carcinoma reveals MGMT as a novel candidate gene in Belarusian children exposed to radiation. BMC Cancer 2017, 17, 328. [Google Scholar] [CrossRef]
- Halkova, T.; Dvorakova, S.; Sykorova, V.; Vaclavikova, E.; Vcelak, J.; Vlcek, P.; Sykorova, P.; Kodetova, D.; Betka, J.; Lastuvka, P.; et al. Polymorphisms in selected DNA repair genes and cell cycle regulating genes involved in the risk of papillary thyroid carcinoma. Cancer Biomark. 2016, 17, 97–106. [Google Scholar] [CrossRef]
- Cipollini, M.; Figlioli, G.; Maccari, G.; Garritano, S.; De Santi, C.; Melaiu, O.; Barone, E.; Bambi, F.; Ermini, S.; Pellegrini, G.; et al. Polymorphisms within base and nucleotide excision repair pathways and risk of differentiated thyroid carcinoma. DNA Repair 2016, 41, 27–31. [Google Scholar] [CrossRef][Green Version]
- Landa, I.; Boullosa, C.; Inglada-Pérez, L.; Sastre-Perona, A.; Pastor, S.; Velázquez, A.; Mancikova, V.; Ruiz-Llorente, S.; Schiavi, F.; Marcos, R.; et al. An Epistatic Interaction between the PAX8 and STK17B Genes in Papillary Thyroid Cancer Susceptibility. PLoS ONE 2013, 8, e74765. [Google Scholar] [CrossRef]
- Liyanarachchi, S.; Wojcicka, A.; Li, W.; Czetwertynska, M.; Stachlewska, E.; Nagy, R.; Hoag, K.; Wen, B.; Ploski, R.; Ringel, M.D.; et al. Cumulative Risk Impact of Five Genetic Variants Associated with Papillary Thyroid Carcinoma. Thyroid 2013, 23, 1532–1540. [Google Scholar] [CrossRef]
- Isakova, J.; Talaibekova, E.; Aldasheva, N.; Vinnikov, D.; Aldashev, A. The association of polymorphic markers Arg399Gln of XRCC1 gene, Arg72Pro of TP53 gene and T309G of MDM2 gene with breast cancer in Kyrgyz females. BMC Cancer 2017, 17, 758. [Google Scholar] [CrossRef]
- Mordukhovich, I.; Beyea, J.; Herring, A.H.; Hatch, M.; Stellman, S.D.; Teitelbaum, S.L.; Richardson, D.B.; Millikan, R.C.; Engel, L.S.; Shantakumar, S.; et al. Polymorphisms in DNA repair genes, traffic-related polycyclic aromatic hydrocarbon exposure and breast cancer incidence. Int. J. Cancer 2016, 139, 310–321. [Google Scholar] [CrossRef]
- Sapkota, Y.; Mackey, J.R.; Lai, R.; Franco-Villalobos, C.; Lupichuk, S.; Robson, P.J.; Kopciuk, K.; Cass, C.E.; Yasui, Y.; Damaraju, S. Assessing SNP-SNP Interactions among DNA Repair, Modification and Metabolism Related Pathway Genes in Breast Cancer Susceptibility. PLoS ONE 2013, 8, e64896. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Shakeri, M.; Zakeri, F.; Changizi, V.; Rajabpour, M.R.; Farshidpour, M.R. Cytogenetic effects of radiation and genetic polymorphisms of the XRCC1 and XRCC3 repair genes in industrial radiographers. Radiat. Environ. Biophys. 2019, 58, 247–255. [Google Scholar] [CrossRef]
- Song, Y.-Z.; Han, F.-J.; Liu, M.; Xia, C.-C.; Shi, W.-Y.; Dong, L.-H. Association between Single Nucleotide Polymorphisms in XRCC3 and Radiation-Induced Adverse Effects on Normal Tissue: A Meta-Analysis. PLoS ONE 2015, 10, e0130388. [Google Scholar] [CrossRef]
- Chai, F.; Liang, Y.; Chen, L.; Zhang, F.; Jiang, J. Association between XRCC3 Thr241Met Polymorphism and Risk of Breast Cancer: Meta-Analysis of 23 Case-Control Studies. Med. Sci. Monit. 2015, 21, 3231–3240. [Google Scholar] [CrossRef]
- Zhang, B.; Beeghly-Fadiel, A.; Long, J.; Zheng, W. Genetic variants associated with breast-cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011, 12, 477–488. [Google Scholar] [CrossRef]
- He, X.-F.; Wei, W.; Li, J.-L.; Shen, X.-L.; Ding, D.-P.; Wang, S.-L.; Liu, Z.-Z.; Qin, J.-B.; Wu, L.-X.; Xie, D.-L. Association between the XRCC3 T241M polymorphism and risk of cancer: Evidence from 157 case–control studies. Gene 2013, 523, 10–19. [Google Scholar] [CrossRef]
- Peng, Q.; Mo, C.; Tang, W.; Chen, Z.; Li, R.; Zhai, L.; Yang, S.; Wu, J.; Sui, J.; Li, S.; et al. DNA repair gene XRCC3 polymorphisms and bladder cancer risk: A meta-analysis. Tumor Biol. 2014, 35, 1933–1944. [Google Scholar] [CrossRef]
- Ma, Q.; Zhao, Y.; Wang, S.; Zhang, X.; Zhang, J.; Du, M.; Li, L.; Zhang, Y. Genetic polymorphisms of XRCC3 Thr241Met (C18067T, rs861539) and bladder cancer risk: A meta-analysis of 18 research studies. Tumor Biol. 2014, 35, 1473–1480. [Google Scholar] [CrossRef]
- Yan, L.; Li, Q.; Li, X.; Ji, H.; Zhang, L. Association Studies between XRCC1, XRCC2, XRCC3 Polymorphisms and Differentiated Thyroid Carcinoma. Cell Physiol. Biochem. 2016, 38, 1075–1084. [Google Scholar] [CrossRef]
- Yuan, K.; Huo, M.; Sun, Y.; Wu, H.; Chen, H.; Wang, Y.; Fu, R. Association between x-ray repair cross-complementing group 3 (XRCC3) genetic polymorphisms and papillary thyroid cancer susceptibility in a Chinese Han population. Tumor Biol. 2016, 37, 979–987. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, K.; Liu, X.; Liu, B.; Wang, Z. Association between XRCC1 and XRCC3 gene polymorphisms and risk of thyroid cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 3160–3167. [Google Scholar]
- Fayaz, S.; Karimmirza, M.; Tanhaei, S.; Fathi, M.; Torbati, P.M.; Fard-Esfahani, P. Increased risk of differentiated thyroid carcinoma with combined effects of homologous recombination repair gene polymorphisms in an Iranian population. Asian Pac. J. Cancer Prev. 2014, 14, 6727–6731. [Google Scholar] [CrossRef]
- Sturgis, E.M.; Zhao, C.; Zheng, R.; Wei, Q. Radiation Response Genotype and Risk of Differentiated Thyroid Cancer: A Case-Control Analysis. Laryngoscope 2005, 115, 938–945. [Google Scholar] [CrossRef]
- Lu, W.; Wu, G.; Zhang, B. Association Between X-Ray Cross-complementing Group 3 (XRCC3) Thr241Met Polymorphism and Risk of Thyroid Cancer: A Meta-Analysis. Med. Sci. Monit. 2015, 21, 3978–3985. [Google Scholar] [CrossRef]
- Yu, X.-L.; Liu, H.; Wang, B.; Fu, Z.-J.; Yuan, Y.; Yan, S.-L.; Zhao, W.-J.; Wang, Y.-G.; Cai, J. Significant associations between X-ray repair cross-complementing group 3 genetic polymorphisms and thyroid cancer risk. Tumor Biol. 2014, 35, 2009–2015. [Google Scholar] [CrossRef]
- Natallia, M.A.; Vladimir, A.S.; Tatiana, I.R.; Valentina, M.D.; Eugeny, F.L.; Victor, K.I.; Norisato, M.; Ryo, K.; Shunichi, Y. Polymorphisms of DNA damage response genes in radiation-related and sporadic papillary thyroid carcinoma. Endocr. Relat. Cancer 2009, 16, 491–503. [Google Scholar] [CrossRef]
- Siraj, A.K.; Al-Rasheed, M.; Ibrahim, M.; Siddiqui, K.; Al-Dayel, F.; Al-Sanea, O.; Uddin, S.; Al-Kuraya, K. RAD52 polymorphisms contribute to the development of papillary thyroid cancer susceptibility in Middle Eastern population. J. Endocrinol. Investig. 2008, 31, 893–899. [Google Scholar] [CrossRef]
- Nouspikel, T. DNA Repair in Mammalian Cells. Cell Mol. Life Sci. 2009, 66, 994–1009. [Google Scholar] [CrossRef]
- Melis, J.P.M.; Luijten, M.; Mullenders, L.H.F.; van Steeg, H. The role of XPC: Implications in cancer and oxidative DNA damage. Mutat. Res. Rev. Mutat. 2011, 728, 107–117. [Google Scholar] [CrossRef]
- Yao, J.-G.; Huang, X.-Y.; Long, X.-D. Interaction of DNA repair gene polymorphisms and aflatoxin B1 in the risk of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 6231–6244. [Google Scholar]
- Chen, Z.; Yang, J.; Wang, G.; Song, B.; Li, J.; Xu, Z. Attenuated Expression of Xeroderma Pigmentosum Group C Is Associated with Critical Events in Human Bladder Cancer Carcinogenesis and Progression. Cancer Res. 2007, 67, 4578–4585. [Google Scholar] [CrossRef]
- Park, J.Y.; Huang, Y.; Sellers, T.A. Single Nucleotide Polymorphisms in DNA Repair Genes and Prostate Cancer Risk. In Cancer Epidemiology. Methods in Molecular Biology; Verma, M., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 471, pp. 361–385. [Google Scholar] [CrossRef]
- Mei, C.-R.; Luo, M.; Li, H.-M.; Deng, W.-J.; Zhou, Q.-H. DNA repair gene polymorphisms in the nucleotide excision repair pathway and lung cancer risk: A meta-analysis. Chin. J. Cancer Res. 2011, 23, 79. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Zhang, W.; Gong, S. An association between XPC Lys939Gln polymorphism and the risk of bladder cancer: A meta-analysis. Tumor Biol. 2013, 34, 973–982. [Google Scholar] [CrossRef]
- Hua, R.-X.; Zhu, J.; Jiang, D.-H.; Zhang, S.-D.; Zhang, J.-B.; Xue, W.-Q.; Li, X.-Z.; Zhang, P.-F.; He, J.; Jia, W.-H. Association of XPC Gene Polymorphisms with Colorectal Cancer Risk in a Southern Chinese Population: A Case-Control Study and Meta-Analysis. Genes 2016, 7, 73. [Google Scholar] [CrossRef]
- He, J.; Shi, T.-Y.; Zhu, M.-L.; Wang, M.-Y.; Li, Q.-X.; Wei, Q.-Y. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: A meta-analysis. Int. J. Cancer 2013, 133, 1765–1775. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, Z.; Shi, X.; Wang, Z. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur. J. Cancer 2008, 44, 2241–2253. [Google Scholar] [CrossRef]
- Francisco, G.; Menezes, P.R.; Eluf-Neto, J.; Chammas, R. XPC polymorphisms play a role in tissue-specific carcinogenesis: A meta-analysis. Eur. J. Hum. Genet. 2008, 16, 724. [Google Scholar] [CrossRef]
- Jin, B.; Dong, Y.; Zhang, X.; Wang, H.; Han, B. Association of XPC Polymorphisms and Lung Cancer Risk: A Meta-Analysis. PLoS ONE 2014, 9, e93937. [Google Scholar] [CrossRef]
- Zhu, M.-L.; Hua, R.-X.; Zheng, L. Associations between polymorphisms of the XPC gene and lung cancer susceptibility: A meta-analysis. Tumor Biol. 2014, 35, 2931–2939. [Google Scholar] [CrossRef]
- Dai, Q.-S.; Hua, R.-X.; Zeng, R.-F.; Long, J.-T.; Peng, Z.-W. XPC gene polymorphisms contribute to bladder cancer susceptibility: A meta-analysis. Tumor Biol. 2014, 35, 447–453. [Google Scholar] [CrossRef]
- Dou, K.; Xu, Q.; Han, X. The association between XPC Lys939Gln gene polymorphism and urinary bladder cancer susceptibility: A systematic review and meta-analysis. Diagn. Pathol. 2013, 8, 112. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, B.; Zheng, W. Genetic variants associated with colorectal cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut 2014, 63, 326–336. [Google Scholar] [CrossRef]
- Peng, Q.; Lao, X.; Tang, W.; Chen, Z.; Li, R.; Qin, X.; Li, S. XPC Lys939Gln polymorphism contributes to colorectal cancer susceptibility: Evidence from a meta-analysis. Diagn. Pathol. 2014, 9, 120. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, L.-T.; Zhang, S.-C.; Chen, K. XPC Polymorphism Increases Risk of Digestive System Cancers: Current Evidence from A Meta-Analysis. Chin. J. Cancer Res. 2012, 24, 181–189. [Google Scholar] [CrossRef][Green Version]
- Long, X.-D.; Ma, Y.; Zhou, Y.-F.; Ma, A.-M.; Fu, G.-H. Polymorphism in xeroderma pigmentosum complementation group C codon 939 and aflatoxin B1–related hepatocellular carcinoma in the Guangxi population. Hepatology 2010, 52, 1301–1309. [Google Scholar] [CrossRef]
- Wang, C.; Nie, H.; Li, Y.; Liu, G.; Wang, X.; Xing, S.; Zhang, L.; Chen, X.; Chen, Y.; Li, Y. The study of the relation of DNA repair pathway genes SNPs and the sensitivity to radiotherapy and chemotherapy of NSCLC. Sci. Rep. 2016, 6, 26526. [Google Scholar] [CrossRef]
- Caronia, D.; Patiño-García, A.; Milne, R.L.; Zalacain-Díez, M.; Pita, G.; Alonso, M.R.; Moreno, L.T.; Sierrasesumaga-Ariznabarreta, L.; Benítez, J.; González-Neira, A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenom. J. 2009, 9, 347. [Google Scholar] [CrossRef]
- Sakano, S.; Hinoda, Y.; Sasaki, M.; Wada, T.; Matsumoto, H.; Eguchi, S.; Shinohara, A.; Kawai, Y.; Hara, T.; Nagao, K.; et al. Nucleotide excision repair gene polymorphisms may predict acute toxicity in patients treated with chemoradiotherapy for bladder cancer. Pharmacogenomics 2010, 11, 1377–1387. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, H.; Chen, Q.; Lin, J.; Grossman, H.B.; Dinney, C.P.; Wu, X.; Gu, J. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair 2008, 7, 141–148. [Google Scholar] [CrossRef][Green Version]
- Pavanello, S.; Pulliero, A.; Siwinska, E.; Mielzynska, D.; Clonfero, E. Reduced nucleotide excision repair and GSTM1-null genotypes influence anti-B[a]PDE–DNA adduct levels in mononuclear white blood cells of highly PAH-exposed coke oven workers. Carcinogenesis 2005, 26, 169–175. [Google Scholar] [CrossRef]
- Lin, J.; Swan, G.E.; Shields, P.G.; Benowitz, N.L.; Gu, J.; Amos, C.I.; de Andrade, M.; Spitz, M.R.; Wu, X. Mutagen Sensitivity and Genetic Variants in Nucleotide Excision Repair Pathway: Genotype-Phenotype Correlation. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2065–2071. [Google Scholar] [CrossRef][Green Version]
- Cornetta, T.; Festa, F.; Testa, A.; Cozzi, R. DNA damage repair and genetic polymorphisms: Assessment of individual sensitivity and repair capacity. Int. J. Radiat. Oncol. 2006, 66, 537–545. [Google Scholar] [CrossRef]
- Qiao, Y.; Spitz, M.R.; Shen, H.; Guo, Z.; Shete, S.; Hedayati, M.; Grossman, L.; Mohrenweiser, H.; Wei, Q. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis 2002, 23, 295–299. [Google Scholar] [CrossRef]
- Ho, V.; Brunetti, V.; Peacock, S.; Massey, T.E.; Godschalk, R.W.L.; van Schooten, F.J.; Ashbury, J.E.; Vanner, S.J.; King, W.D. Exposure to meat-derived carcinogens and bulky DNA adduct levels in normal-appearing colon mucosa. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2017, 821, 5–12. [Google Scholar] [CrossRef]
- Broaddus, R.R.; Lynch, P.M.; Lu, K.H.; Luthra, R.; Michelson, S.J. Unusual tumors associated with the hereditary nonpolyposis colorectal cancer syndrome. Mod. Pathol. 2004, 17, 981–989. [Google Scholar] [CrossRef][Green Version]
- Pande, M.; Wei, C.; Chen, J.; Amos, C.I.; Lynch, P.M.; Lu, K.H.; Lucio, L.A.; Boyd-Rogers, S.G.; Bannon, S.A.; Mork, M.E.; et al. Cancer spectrum in DNA mismatch repair gene mutation carriers: Results from a hospital based Lynch syndrome registry. Fam. Cancer 2012, 11, 441–447. [Google Scholar] [CrossRef]
- Stulp, R.P.; Herkert, J.C.; Karrenbeld, A.; Mol, B.; Vos, Y.J.; Sijmons, R.H. Thyroid cancer in a patient with a germline MSH2 mutation. Case report and review of the Lynch syndrome expanding tumour spectrum. Hered. Cancer Clin. Pract. 2008, 6, 15–21. [Google Scholar] [CrossRef]
- Pelizzo, M.R.; Pennelli, G.; Zane, M.; Galuppini, F.; Colletti, P.M.; Merante Boschin, I.; Rubello, D. Papillary thyroid carcinoma (PTC) in Lynch syndrome: Report of two cases and discussion on Lynch syndrome behaviour and genetics. BioMed Pharmacother. 2015, 74, 9–16. [Google Scholar] [CrossRef]
- Johnson, J.M.; Chen, J.; Ali, S.M.; Dardi, I.K.; Tuluc, M.; Cognetti, D.; Campling, B.; Sama, A.R. Molecular Profiling of Synchronous Colon Cancers and Anaplastic Thyroid Cancer in a Patient with Lynch Syndrome. J. Gastrointest. Cancer 2018, 49, 203–206. [Google Scholar] [CrossRef]
- Kunstman, J.W.; Juhlin, C.C.; Goh, G.; Brown, T.C.; Stenman, A.; Healy, J.M.; Rubinstein, J.C.; Choi, M.; Kiss, N.; Nelson-Williams, C.; et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum. Mol. Genet. 2015, 24, 2318–2329. [Google Scholar] [CrossRef]
- Yu, Y.; Dong, L.; Li, D.; Chuai, S.; Wu, Z.; Zheng, X.; Cheng, Y.; Han, L.; Yu, J.; Gao, M. Targeted DNA Sequencing Detects Mutations Related to Susceptibility among Familial Non-medullary Thyroid Cancer. Sci. Rep. 2015, 5, 16129. [Google Scholar] [CrossRef]
- Picelli, S.; Zajac, P.; Zhou, X.-L.; Edler, D.; Lenander, C.; Dalén, J.; Hjern, F.; Lundqvist, N.; Lindforss, U.; Påhlman, L.; et al. Common variants in human CRC genes as low-risk alleles. Eur. J. Cancer 2010, 46, 1041–1048. [Google Scholar] [CrossRef]
- Giráldez, M.D.; Balaguer, F.; Caldés, T.; Sanchez-de-Abajo, A.; Gómez-Fernández, N.; Ruiz-Ponte, C.; Muñoz, J.; Garre, P.; Gonzalo, V.; Moreira, L.; et al. Association of MUTYH and MSH6 germline mutations in colorectal cancer patients. Fam. Cancer 2009, 8, 525. [Google Scholar] [CrossRef]
- Conde, J.; Silva, S.N.; Azevedo, A.P.; Teixeira, V.; Pina, J.E.; Rueff, J.; Gaspar, J.F. Association of common variants in mismatch repair genes and breast cancer susceptibility: A multigene study. BMC Cancer 2009, 9, 344. [Google Scholar] [CrossRef]
- Curtin, K.; Samowitz, W.S.; Wolff, R.K.; Caan, B.J.; Ulrich, C.M.; Potter, J.D.; Slattery, M.L. MSH6 G39E polymorphism and CpG island methylator phenotype in colon cancer. Mol. Carcinog. 2009, 48, 989–994. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Kawamoto, K.; Kikuno, N.; Suehiro, Y.; Okayama, N.; Tanaka, Y.; Dahiya, R. Mismatch Repair Gene MSH3 Polymorphism is Associated with the Risk of Sporadic Prostate Cancer. J. Urol. 2008, 179, 2020–2024. [Google Scholar] [CrossRef]
- Smith, T.R.; Levine, E.A.; Freimanis, R.I.; Akman, S.A.; Allen, G.O.; Hoang, K.N.; Liu-Mares, W.; Hu, J.J. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 2008, 29, 2132–2138. [Google Scholar] [CrossRef]
- Landi, S.; Gemignani, F.; Canzian, F.; Gaborieau, V.; Barale, R.; Landi, D.; Szeszenia-Dabrowska, N.; Zaridze, D.; Lissowska, J.; Rudnai, P.; et al. DNA Repair and Cell Cycle Control Genes and the Risk of Young-Onset Lung Cancer. Cancer Res. 2006, 66, 11062–11069. [Google Scholar] [CrossRef][Green Version]
- Yu, J.H.; Bigler, J.; Whitton, J.; Potter, J.D.; Ulrich, C.M. Mismatch repair polymorphisms and colorectal polyps: hMLH1-93G>A variant modifies risk associated with smoking. Am. J. Gastroenterol. 2006, 101, 1313–1319. [Google Scholar] [CrossRef]
- Zelga, P.; Przybyłowska-Sygut, K.; Zelga, M.; Dziki, A.; Majsterek, I. The 116G > A MSH6 and IVS1-1121C > T PMS2 Genes Polymorphisms Modulate the Risk of the Sporadic Colorectal Cancer Development in Polish Population. Pathol. Oncol. Res. 2018, 24, 231–235. [Google Scholar] [CrossRef]
- Berndt, S.I.; Platz, E.A.; Fallin, M.D.; Thuita, L.W.; Hoffman, S.C.; Helzlsouer, K.J. Mismatch repair polymorphisms and the risk of colorectal cancer. Int. J. Cancer 2007, 120, 1548–1554. [Google Scholar] [CrossRef]
- Campbell, P.T.; Curtin, K.; Ulrich, C.M.; Samowitz, W.S.; Bigler, J.; Velicer, C.M.; Caan, B.; Potter, J.D.; Slattery, M.L. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut 2009, 58, 661–667. [Google Scholar] [CrossRef]
- Sanyal, S.; De Verdier, P.J.; Steineck, G.; Larsson, P.; Onelov, E.; Hemminki, K.; Kumar, R. Polymorphisms in XPD, XPC and the risk of death in patients with urinary bladder neoplasms. Acta Oncol. 2007, 46, 31–41. [Google Scholar] [CrossRef]
- Dong, X.; Li, Y.; Chang, P.; Hess, K.R.; Abbruzzese, J.L.; Li, D. DNA mismatch repair network gene polymorphism as a susceptibility factor for pancreatic cancer. Mol. Carcinog. 2012, 51, 491–499. [Google Scholar] [CrossRef]
- Lee, E.; Levine, E.A.; Franco, V.I.; Allen, G.O.; Gong, F.; Zhang, Y.; Hu, J.J. Combined Genetic and Nutritional Risk Models of Triple Negative Breast Cancer. Nutr. Cancer 2014, 66, 955–963. [Google Scholar] [CrossRef]
- Tulupova, E.; Kumar, R.; Hanova, M.; Slyskova, J.; Pardini, B.; Polakova, V.; Naccarati, A.; Vodickova, L.; Novotny, J.; Halamkova, J.; et al. Do polymorphisms and haplotypes of mismatch repair genes modulate risk of sporadic colorectal cancer? Mutat. Res. 2008, 648, 40–45. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Jia, J.; Tang, L.; Gao, X.; Yan, L.; Wang, L.; Yu, F.; Ma, N.; Liu, W.; et al. Correlation between polymorphisms in DNA mismatch repair genes and the risk of primary hepatocellular carcinoma for the Han population in northern China. Scand. J. Gastroenterol. 2015, 50, 1404–1410. [Google Scholar] [CrossRef]
- Li, Z.; Kong, L.; Yu, L.; Huang, J.; Wang, K.; Chen, S.; Yu, M.; Wei, S. Association between MSH6 G39E polymorphism and cancer susceptibility: A meta-analysis of 7,046 cases and 34,554 controls. Tumour Biol. 2014, 35, 6029–6037. [Google Scholar] [CrossRef]
- Chen, M.; Kamat, A.M.; Huang, M.; Grossman, H.B.; Dinney, C.P.; Lerner, S.P.; Wu, X.; Gu, J. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis 2007, 28, 2160–2165. [Google Scholar] [CrossRef][Green Version]
- Enjuanes, A.; Benavente, Y.; Bosch, F.; Martín-Guerrero, I.; Colomer, D.; Pérez-Álvarez, S.; Reina, O.; Ardanaz, M.T.; Jares, P.; García-Orad, A.; et al. Genetic Variants in Apoptosis and Immunoregulation-Related Genes Are Associated with Risk of Chronic Lymphocytic Leukemia. Cancer Res. 2008, 68, 10178–10186. [Google Scholar] [CrossRef]
- Pan, J.; Lin, J.; Izzo, J.G.; Liu, Y.; Xing, J.; Huang, M.; Ajani, J.A.; Wu, X. Genetic susceptibility to esophageal cancer: The role of the nucleotide excision repair pathway. Carcinogenesis 2009, 30, 785–792. [Google Scholar] [CrossRef]
- Wu, X.; Gu, J.; Grossman, H.B.; Amos, C.I.; Etzel, C.; Huang, M.; Zhang, Q.; Millikan, R.E.; Lerner, S.; Dinney, C.P.; et al. Bladder Cancer Predisposition: A Multigenic Approach to DNA-Repair and Cell-Cycle–Control Genes. Am. J. Hum. Genet. 2006, 78, 464–479. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, W.-Y.; Andreotti, G.; Gao, Y.-T.; Rashid, A.; Chen, J.; Sakoda, L.C.; Shen, M.-C.; Wang, B.-S.; Chanock, S.; et al. Variants of DNA Repair Genes and the Risk of Biliary Tract Cancers and Stones: A Population-Based Study in China. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2123–2127. [Google Scholar] [CrossRef]
- Lin, J.; Pu, X.; Wang, W.; Matin, S.; Tannir, N.M.; Wood, C.G.; Wu, X. Case–control analysis of nucleotide excision repair pathway and the risk of renal cell carcinoma. Carcinogenesis 2008, 29, 2112–2119. [Google Scholar] [CrossRef]
- Wang, Y.; Spitz, M.R.; Lee, J.J.; Huang, M.; Lippman, S.M.; Wu, X. Nucleotide Excision Repair Pathway Genes and Oral Premalignant Lesions. Clin. Cancer Res. 2007, 13, 3753–3758. [Google Scholar] [CrossRef]
- Wu, X.; Lu, C.; Ye, Y.; Chang, J.; Yang, H.; Lin, J.; Gu, J.; Hong, W.K.; Stewart, D.; Spitz, M.R. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet. Genom. 2008, 18, 955–965. [Google Scholar] [CrossRef]
- Palugulla, S.; Devaraju, P.; Kayal, S.; Narayan, S.K.; Mathaiyan, J. Genetic polymorphisms in cyclin H gene are associated with oxaliplatin-induced acute peripheral neuropathy in South Indian digestive tract cancer patients. Cancer Chemother. Pharm. 2018, 82, 421–428. [Google Scholar] [CrossRef]
- Custodio, A.; Moreno-Rubio, J.; Aparicio, J.; Gallego-Plazas, J.; Yaya, R.; Maurel, J.; Higuera, O.; Burgos, E.; Ramos, D.; Calatrava, A.; et al. Pharmacogenetic predictors of severe peripheral neuropathy in colon cancer patients treated with oxaliplatin-based adjuvant chemotherapy: A GEMCAD group study. Ann. Oncol. 2013, 25, 398–403. [Google Scholar] [CrossRef]
- Lolli, G.; Johnson, L.N. CAK-Cyclin-dependent Activating Kinase: A key kinase in cell cycle control and a target for drugs? Cell cycle 2005, 4, 572–577. [Google Scholar] [CrossRef]
- Emmert, S.; Schneider, T.D.; Khan, S.G.; Kraemer, K.H. The human XPG gene: Gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001, 29, 1443–1452. [Google Scholar] [CrossRef]
- Sang, L.; Lv, Z.; Sun, L.-P.; Xu, Q.; Yuan, Y. Impact of SNP-SNP interactions of DNA repair gene ERCC5 and metabolic gene GSTP1 on gastric cancer/atrophic gastritis risk in a Chinese population. World J. Gastroenterol. 2018, 24, 602–612. [Google Scholar] [CrossRef]
- Negovan, A.; Iancu, M.; Moldovan, V.; Mocan, S.; Banescu, C. The Interaction between GSTT1, GSTM1, and GSTP1 Ile105Val Gene Polymorphisms and Environmental Risk Factors in Premalignant Gastric Lesions Risk. BioMed Res. Int. 2017, 2017, 7365080. [Google Scholar] [CrossRef]
- Khabaz, M.N. Polymorphism of the glutathione S-transferase P1 gene (GST-pi) in breast carcinoma. Pol. J. Pathol. 2014, 65, 141–146. [Google Scholar] [CrossRef]
- Cheng, L.; Sturgis, E.M.; Eicher, S.A.; Spitz, M.R.; Wei, Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer 2002, 94, 393–397. [Google Scholar] [CrossRef]
- Hussain, S.K.; Mu, L.-N.; Cai, L.; Chang, S.-C.; Park, S.L.; Oh, S.S.; Wang, Y.; Goldstein, B.Y.; Ding, B.-G.; Jiang, Q.; et al. Genetic Variation in Immune Regulation and DNA Repair Pathways and Stomach Cancer in China. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2304–2309. [Google Scholar] [CrossRef]
- Paszkowska-Szczur, K.; Scott, R.J.; Serrano-Fernandez, P.; Mirecka, A.; Gapska, P.; Górski, B.; Cybulski, C.; Maleszka, R.; Sulikowski, M.; Nagay, L.; et al. Xeroderma pigmentosum genes and melanoma risk. Int. J. Cancer 2013, 133, 1094–1100. [Google Scholar] [CrossRef]
- Ma, H.; Yu, H.; Liu, Z.; Wang, L.-E.; Sturgis, E.M.; Wei, Q. Polymorphisms of XPG/ERCC5 and risk of squamous cell carcinoma of the head and neck. Pharmacogenet. Genom. 2012, 22, 50–57. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Tang, L.-L.; Long, J.-T.; Zhu, J.; Hua, R.-X.; Li, J. XPG gene polymorphisms and cancer susceptibility: Evidence from 47 studies. Oncotarget 2017, 8, 37263–37277. [Google Scholar] [CrossRef]
- Su, C.-H.; Chang, W.-S.; Hu, P.-S.; Hsiao, C.-L.; Ji, H.-X.; Liao, C.-H.; Yueh, T.-C.; Chuang, C.-L.; Tsai, C.-W.; Hsu, C.-M.; et al. Contribution of DNA Double-strand Break Repair Gene XRCC3 Genotypes to Triple-negative Breast Cancer Risk. Cancer Genom. Proteom. 2015, 12, 359–367. [Google Scholar]
- Derwahl, M.; Nicula, D. Estrogen and its role in thyroid cancer. Endocr. Relat. Cancer 2014, 21, T273–T283. [Google Scholar] [CrossRef]
- Nielsen, S.M.; White, M.G.; Hong, S.; Aschebrook-Kilfoy, B.; Kaplan, E.L.; Angelos, P.; Kulkarni, S.A.; Olopade, O.I.; Grogan, R.H. The Breast–Thyroid Cancer Link: A Systematic Review and Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2016, 25, 231–238. [Google Scholar] [CrossRef]
- Chuffa, L.G.; Lupi-Júnior, L.A.; Costa, A.B.; Amorim, J.P.; Seiva, F.R. The role of sex hormones and steroid receptors on female reproductive cancers. Steroids 2017, 118, 93–108. [Google Scholar] [CrossRef]
- Cavalieri, E.; Rogan, E. The molecular etiology and prevention of estrogen-initiated cancers: Ockham’s Razor: Pluralitas non est ponenda sine necessitate. Plurality should not be posited without necessity. Mol. Asp. Med. 2014, 36, 1–55. [Google Scholar] [CrossRef]
- Vahteristo, P.; Ojala, S.; Tamminen, A.; Tommiska, J.; Sammalkorpi, H.; Kiuru-Kuhlefelt, S.; Eerola, H.; Aaltonen, L.A.; Aittomäki, K.; Nevanlinna, H. No MSH6 germline mutations in breast cancer families with colorectal and/or endometrial cancer. J. Med. Genet. 2005, 42, e22. [Google Scholar] [CrossRef]
- Hsieh, P.; Yamane, K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008, 129, 391–407. [Google Scholar] [CrossRef]
- Peltomäki, P. Update on Lynch syndrome genomics. Fam. Cancer 2016, 15, 385–393. [Google Scholar] [CrossRef]
- Lynch, H.T.; Shaw, T.G. Practical genetics of colorectal cancer. Chin. Clin. Oncol. 2013, 2, 12. [Google Scholar] [CrossRef]
- Wada-Hiraike, O.; Yano, T.; Nei, T.; Matsumoto, Y.; Nagasaka, K.; Takizawa, S.; Oishi, H.; Arimoto, T.; Nakagawa, S.; Yasugi, T.; et al. The DNA mismatch repair gene hMSH2 is a potent coactivator of oestrogen receptor [alpha]. Br. J. Cancer 2005, 92, 2286–2291. [Google Scholar] [CrossRef]
- Dralle, H.; Machens, A.; Basa, J.; Fatourechi, V.; Franceschi, S.; Hay, I.D.; Nikiforov, Y.E.; Pacini, F.; Pasieka, J.L.; Sherman, S.I. Follicular cell-derived thyroid cancer. Nat. Rev. Dis. Prim. 2015, 1, 15077. [Google Scholar] [CrossRef]
- Li, C.; Yin, M.; Wang, L.-E.; Amos, C.I.; Zhu, D.; Lee, J.E.; Gershenwald, J.E.; Grimm, E.A.; Wei, Q. Polymorphisms of Nucleotide Excision Repair Genes Predict Melanoma Survival. J. Investig. Dermatol. 2013, 133, 1813–1821. [Google Scholar] [CrossRef]
- Mumbrekar, K.D.; Goutham, H.V.; Vadhiraja, B.M.; Bola Sadashiva, S.R. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals. DNA Repair 2016, 40, 27–34. [Google Scholar] [CrossRef]
- Edelbrock, M.A.; Kaliyaperumal, S.; Williams, K.J. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat. Res. 2013, 743, 53–66. [Google Scholar] [CrossRef]
- Bridge, G.; Rashid, S.; Martin, S. DNA Mismatch Repair and Oxidative DNA Damage: Implications for Cancer Biology and Treatment. Cancers 2014, 6, 1597–1614. [Google Scholar] [CrossRef]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef]
- Kamiza, A.B.; Hsieh, L.-L.; Tang, R.; Chien, H.-T.; Lai, C.-H.; Chiu, L.-L.; Lo, T.-P.; Hung, K.-Y.; You, J.-F.; Wang, W.-C.; et al. Polymorphisms of DNA repair genes are associated with colorectal cancer in patients with Lynch syndrome. Mol. Genet. Genom. Med. 2018, 6, 533–540. [Google Scholar] [CrossRef]
- Vral, A.; Willems, P.; Claes, K.; Poppe, B.; Perletti, G.; Thierens, H. Combined effect of polymorphisms in Rad51 and Xrcc3 on breast cancer risk and chromosomal radiosensitivity. Mol. Med. Rep. 2011, 4, 901–912. [Google Scholar] [CrossRef]
- Bishop, D.K.; Ear, U.; Bhattacharyya, A.; Calderone, C.; Beckett, M.; Weichselbaum, R.R.; Shinohara, A. Xrcc3 Is Required for Assembly of Rad51 Complexes in vivo. J. Biol. Chem. 1998, 273, 21482–21488. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).