Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of CC-Type GRX Protein-Coding Genes in the Maize Genome
2.2. Gene Structure and Phylogenetic Relationships Analysis
2.3. Identification of Cis-Regulatory Elements in Promoters of ZmGRXCCs
2.4. Expression Profile Analysis
2.4.1. Plant Treatments
2.4.2. RNA Isolation and Real-Time PCR Analysis
2.5. Association Analysis
3. Results
3.1. Forty-Five Maize GRXs, Including Twenty-One CC-Type GRX (ZmGRXCC) Genes, Were Identified
3.2. Phylogenetic Relationship Analysis Showed High Conservation in ZmGRXCC Genes
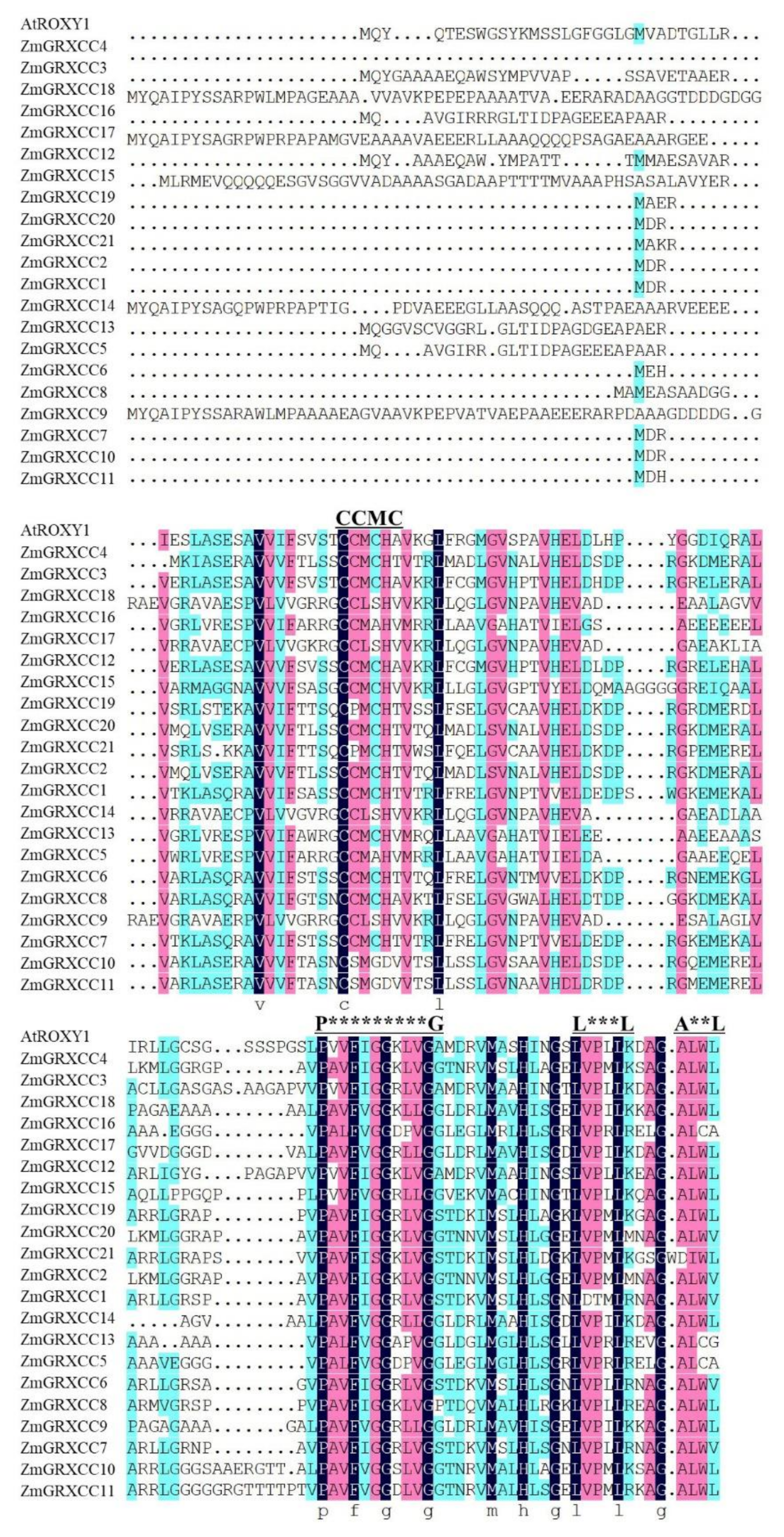
3.3. ZmGRXCC Proteins Were Highly Conserved Containing an Active Site and a C-Terminal ALWL Sequence
3.4. Promoter Analysis of the ZmGRXCC Gene Family
3.5. Expression Profile of ZmGRXCCs under Stress Treatments
3.6. Natural Variations in ZmGRXCC14 Are Associated with Drought Tolerance in Maize
3.7. Expression Profile of ZmGRXCC14 and 17 Are Both Induced by Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mh, C.D.C. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Jacquot, J.-P.; Rouhier, N. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.-P.; Riondet, C. Thioredoxin and Glutaredoxin Systems in Plants: Molecular Mechanisms, Crosstalks, and Functional Significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef]
- Kanda, M.; Ihara, Y.; Murata, H.; Urata, Y.; Kono, T.; Yodoi, J.; Seto, S.; Yano, K.; Kondo, T. Glutaredoxin Modulates Platelet-derived Growth Factor-dependent Cell Signaling by Regulating the Redox Status of Low Molecular Weight Protein-tyrosine Phosphatase. J. Biol. Chem. 2006, 281, 28518–28528. [Google Scholar] [CrossRef]
- Carroll, M.C.; Outten, C.E.; Proescher, J.B.; Rosenfeld, L.; Watson, W.H.; Whitson, L.J.; Hart, P.J.; Jensen, L.T.; Culotta, V.C. The Effects of Glutaredoxin and Copper Activation Pathways on the Disulfide and Stability of Cu,Zn Superoxide Dismutase. J. Biol. Chem. 2006, 281, 28648–28656. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Juszczuk, I.M.; Szal, B.; Rychter, A.M. Oxidation–reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction. Plant Cell Environ. 2012, 35, 296–307. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, S.; Birkenbihl, R.P.; Zachgo, S. Conserved Functions of Arabidopsis and Rice CC-Type Glutaredoxins in Flower Development and Pathogen Response. Mol. Plant 2009, 2, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Ruan, M.-B.; Yang, Y.-L.; Li, K.-M.; Guo, X.; Wang, B.; Yu, X.-L.; Peng, M. Identification and characterization of drought-responsive CC-type glutaredoxins from cassava cultivars reveals their involvement in ABA signalling. BMC Plant Biol. 2018, 18, 329. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-Wide Survey and Expression Analysis Suggest Diverse Roles of Glutaredoxin Gene Family Members during Development and Response to Various Stimuli in Rice. DNA Res. 2010, 17, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Rosso, M.G.; Zachgo, S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 2005, 132, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, M.; Bhave, M.; Zachgo, S. Origin and Diversification of Land Plant CC-Type Glutaredoxins. Genome Biol. Evol. 2009, 1, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Didierjean, C.; Jacquot, J.-P.; Rouhier, N. Engineered mutated glutaredoxins mimicking peculiar plant class III glutaredoxins bind iron–sulfur centers and possess reductase activity. Biochem. Biophys. Res. Commun. 2010, 403, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008, 53, 790–801. [Google Scholar] [CrossRef]
- L’Haridon, F.; Astier, J.; Zander, M.; Page, G.; Thurow, C.; Wendehenne, D.; Gatz, C.; Métraux, J.-P.; Abou-Mansour, E.; La Camera, S.; et al. The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 2011, 68, 507–519. [Google Scholar]
- Laporte, D.; Olate, E.; Salinas, P.; Salazar, M.; Jordana, X.; Holuigue, L. Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Exp. Bot. 2012, 63, 503–515. [Google Scholar] [CrossRef]
- Hong, L.; Tang, D.; Zhu, K.; Wang, K.; Li, M.; Cheng, Z. Somatic and Reproductive Cell Development in Rice Anther Is Regulated by a Putative Glutaredoxin. Plant Cell 2012, 24, 577–588. [Google Scholar] [CrossRef]
- Sharma, R.; Priya, P.; Jain, M. Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta 2013, 238, 871–884. [Google Scholar] [CrossRef] [PubMed]
- El-Kereamy, A.; Bi, Y.-M.; Mahmood, K.; Ranathunge, K.; Yaish, M.W.; Nambara, E.; Rothstein, S.J. Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front. Plant Sci. 2015, 6, 934. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, N.; Thurow, C.; Zachgo, S.; Gatz, C. Plant-specific CC-type glutaredoxins: Functions in developmental processes and stress responses. Biol. Chem. 2015, 396, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Yu, C. China’s water crisis needs more than words. Nature 2011, 470, 307. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qin, F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017, 8, 1110. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Marchlerbauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; Deweesescott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; Mcgettigan, P.; Mc William, H.; Valentin, F.; Wallace, I.; Wilm, A.; López, R.; et al. Clustal W and Clustal X version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.; Yoshiwara, K. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Mathiyalagan, R.; Muthurajan, R.; Subramaniyam, S.; Jegadeesan, R. Insilico analysis of drought tolerant genes in rice. Int. J. Biol. Med Res. 2010, 1, 36–40. [Google Scholar]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.W.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; De Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Wang, H.; Xin, H.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013, 9, e1003790. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Feng, X.; Du, H.; Wang, H. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ 2019, 7, e6765. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Hobo, T.; Asada, M.; Kowyama, Y.; Hattori, T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999, 19, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yang, J.; Cheng, N.; Hirschi, K.D.; White, F.F.; Park, S. Glutaredoxins in plant development, abiotic stress response, and iron homeostasis: From model organisms to crops. Environ. Exp. Bot. 2017, 139, 91–98. [Google Scholar] [CrossRef]






| Identifier | Gene Symbol | Chromosome | Class | Redox Site | ALWL-Motif |
|---|---|---|---|---|---|
| GRMZM2G413315 | ZmGRXCC1 | 1 | CC | CCMC | ALWV |
| GRMZM2G469994 | ZmGRXCC2 | 1 | CC | CCMC | ALWV |
| GRMZM2G480903 | ZmGRXCC3 | 2 | CC | CCMC | ALWL |
| GRMZM5G892308 | ZmGRXCC4 | 2 | CC | CCMC | ALWL |
| GRMZM2G303044 | ZmGRXCC5 | 3 | CC | CCMA | ALCA |
| GRMZM2G110286 | ZmGRXCC6 | 3 | CC | CCMC | ALWV |
| GRMZM2G052796 | ZmGRXCC7 | 3 | CC | CCMC | ALWV |
| GRMZM2G403680 | ZmGRXCC8 | 3 | CC | CCMC | ALWL |
| GRMZM2G023237 | ZmGRXCC9 | 3 | CC | CCLS | ALWL |
| GRMZM2G371063 | ZmGRXCC10 | 4 | CC | CSMG | ALWL |
| GRMZM5G860607 | ZmGRXCC11 | 4 | CC | CSMG | ALWL |
| GRMZM2G470756 | ZmGRXCC12 | 5 | CC | CCMC | ALWL |
| GRMZM2G318213 | ZmGRXCC13 | 6 | CC | CCMC | ALCG |
| GRMZM2G318180 | ZmGRXCC14 | 6 | CC | CCLS | ALWL |
| GRMZM2G442791 | ZmGRXCC15 | 7 | CC | CCMC | ALWL |
| GRMZM2G441906 | ZmGRXCC16 | 8 | CC | CCMA | ALCA |
| GRMZM2G311898 | ZmGRXCC17 | 8 | CC | CCLS | ALWL |
| GRMZM2G178886 | ZmGRXCC18 | 8 | CC | CCLS | ALWL |
| GRMZM2G337706 | ZmGRXCC19 | 10 | CC | CPMC | AIWL |
| GRMZM2G457898 | ZmGRXCC20 | 10 | CC | CCMC | ALWV |
| GRMZM2G303536 | ZmGRXCC21 | 10 | CC | CPMC | DIWL |
| Locus ID | Gene Name | Polymorphic | GLM | PC2 | MLM | |
|---|---|---|---|---|---|---|
| p ≤ 0.01 | p ≤ 0.01 | p ≤ 0.01 | p ≤ 0.005 | |||
| GRMZM2G413315 | ZmGRXCC1 | 1 | - | - | - | - |
| GRMZM2G469994 | ZmGRXCC2 | - | - | - | - | - |
| GRMZM2G480903 | ZmGRXCC3 | 1 | 1 | - | - | - |
| GRMZM5G892308 | ZmGRXCC4 | - | - | - | - | - |
| GRMZM2G303044 | ZmGRXCC5 | 43 | - | - | - | - |
| GRMZM2G110286 | ZmGRXCC6 | 1 | - | - | - | - |
| GRMZM2G052796 | ZmGRXCC7 | 3 | - | - | - | - |
| GRMZM2G403680 | ZmGRXCC8 | - | - | - | - | - |
| GRMZM2G023237 | ZmGRXCC9 | 23 | - | - | - | - |
| GRMZM2G371063 | ZmGRXCC10 | - | - | - | - | - |
| GRMZM5G860607 | ZmGRXCC11 | - | - | - | - | - |
| GRMZM2G470756 | ZmGRXCC12 | 1 | - | - | - | - |
| GRMZM2G318213 | ZmGRXCC13 | 11 | - | - | - | - |
| GRMZM2G318180 | ZmGRXCC14 | 20 | 10 | 1 | 1 | 1 |
| GRMZM2G442791 | ZmGRXCC15 | - | - | - | - | - |
| GRMZM2G441906 | ZmGRXCC16 | 20 | 4 | - | 1 | - |
| GRMZM2G311898 | ZmGRXCC17 | 1 | - | - | - | - |
| GRMZM2G178886 | ZmGRXCC18 | 34 | 10 | - | - | - |
| GRMZM2G337706 | ZmGRXCC19 | - | - | - | - | - |
| GRMZM2G457898 | ZmGRXCC20 | - | - | - | - | - |
| GRMZM2G303536 | ZmGRXCC21 | - | - | - | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.; He, F.; Tang, W.; Du, H.; Wang, H. Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes 2019, 10, 610. https://doi.org/10.3390/genes10080610
Ding S, He F, Tang W, Du H, Wang H. Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes. 2019; 10(8):610. https://doi.org/10.3390/genes10080610
Chicago/Turabian StyleDing, Shuangcheng, Fengyu He, Wenlin Tang, Hewei Du, and Hongwei Wang. 2019. "Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress" Genes 10, no. 8: 610. https://doi.org/10.3390/genes10080610
APA StyleDing, S., He, F., Tang, W., Du, H., & Wang, H. (2019). Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes, 10(8), 610. https://doi.org/10.3390/genes10080610





