Quantifying Risk Pathway Crosstalk Mediated by miRNA to Screen Precision drugs for Breast Cancer Patients
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Matched miRNA/Gene Expression Profiles and Clinical Data
2.1.1. miRNA-Target Relationship Data
2.1.2. PPI Network and Pathway Data
2.1.3. Drug and Drug Target Data
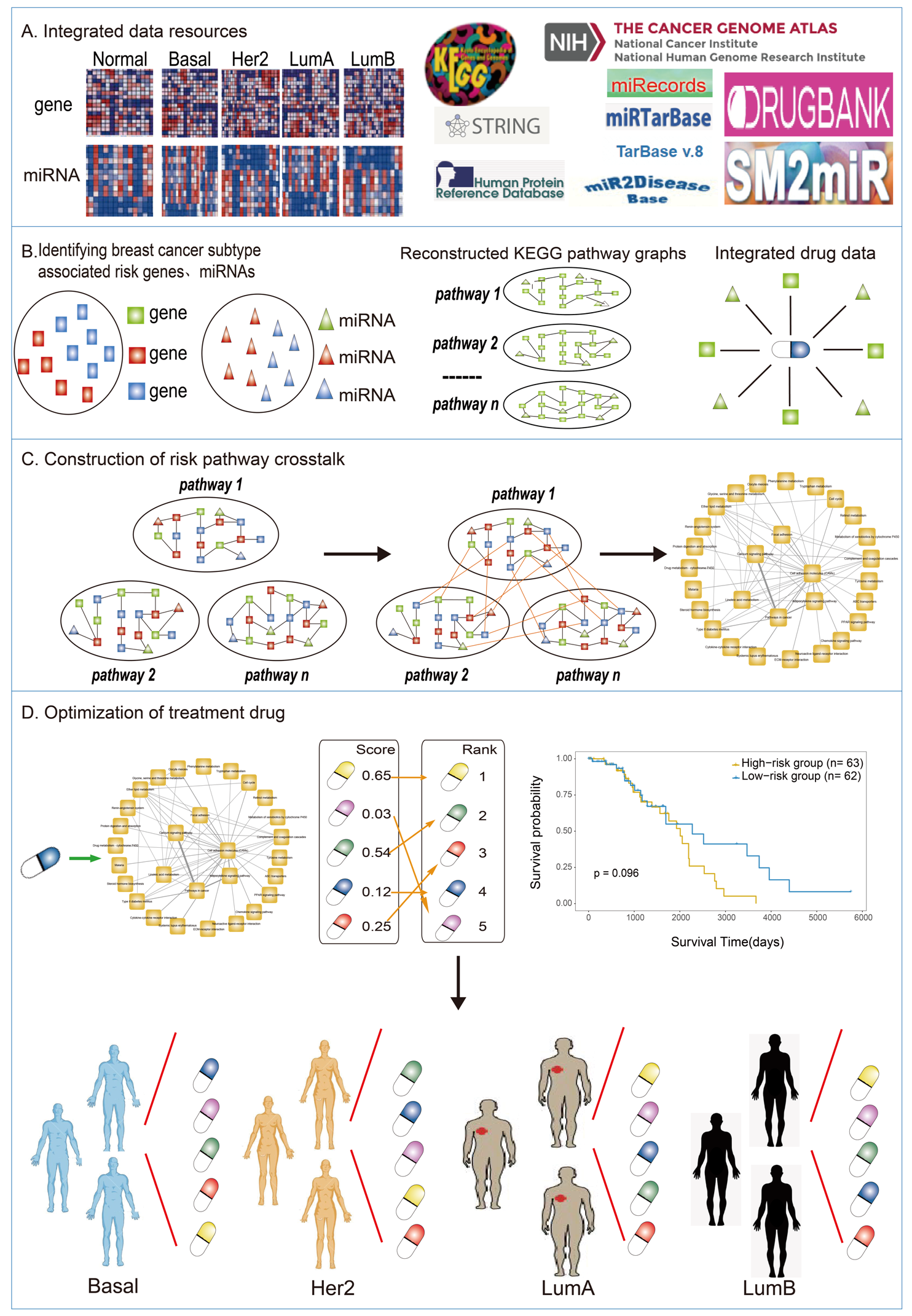
2.2. Reconstructed KEGG Pathway Graphs
2.3. Identification of Risk Genes and miRNAs Related to Breast Cancer Subtypes
2.4. Mining Risk Pathways Associated with Breast Cancer Subtypes
2.5. Establishing the Risk Pathways’ Crosstalk of Breast Cancer
2.6. Evaluating the Impacts of Drugs on Crosstalk
2.7. Survival Analysis
3. Results
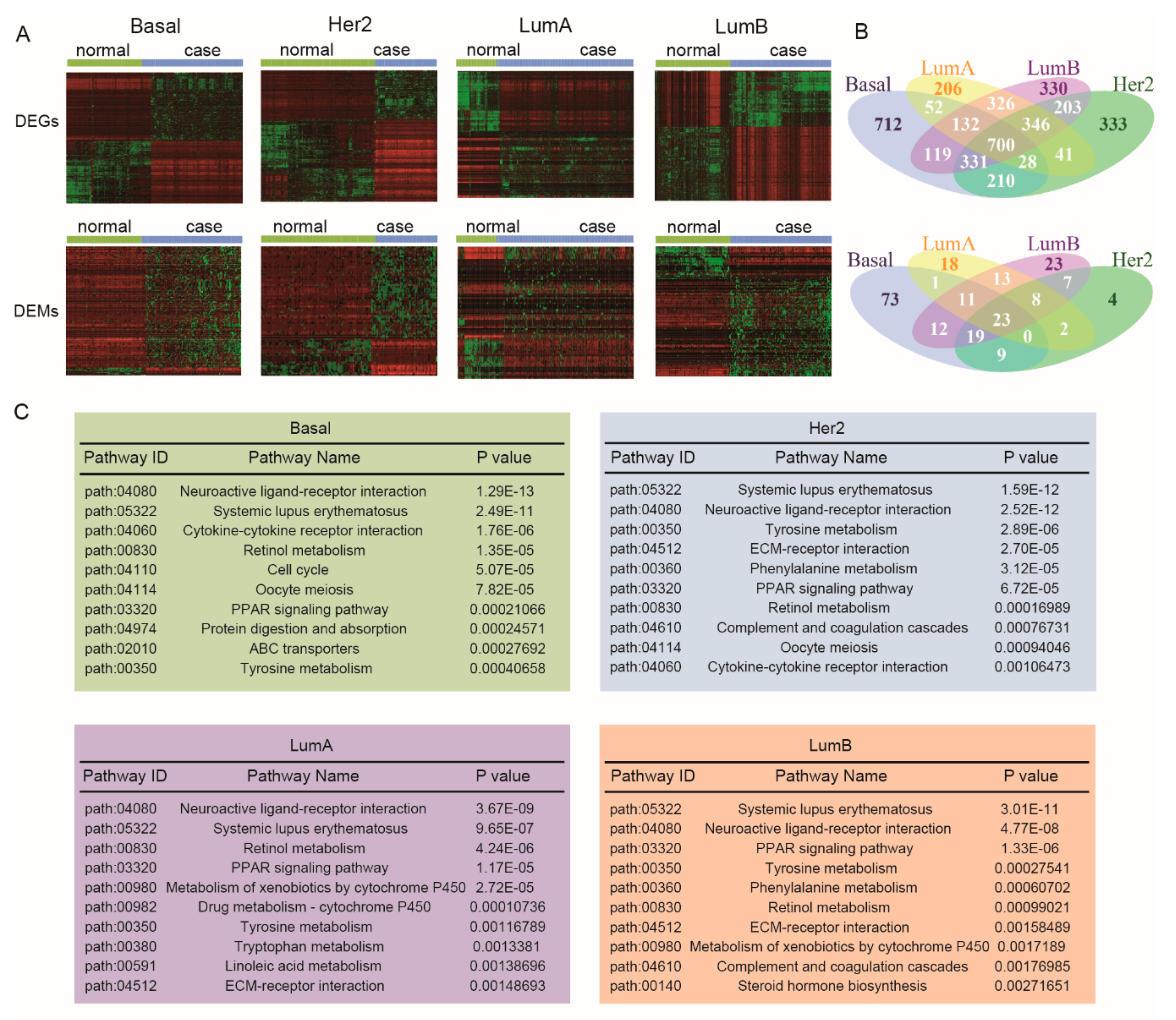
3.1. Identifying Breast Cancer Subtype-Associated Risk Pathways
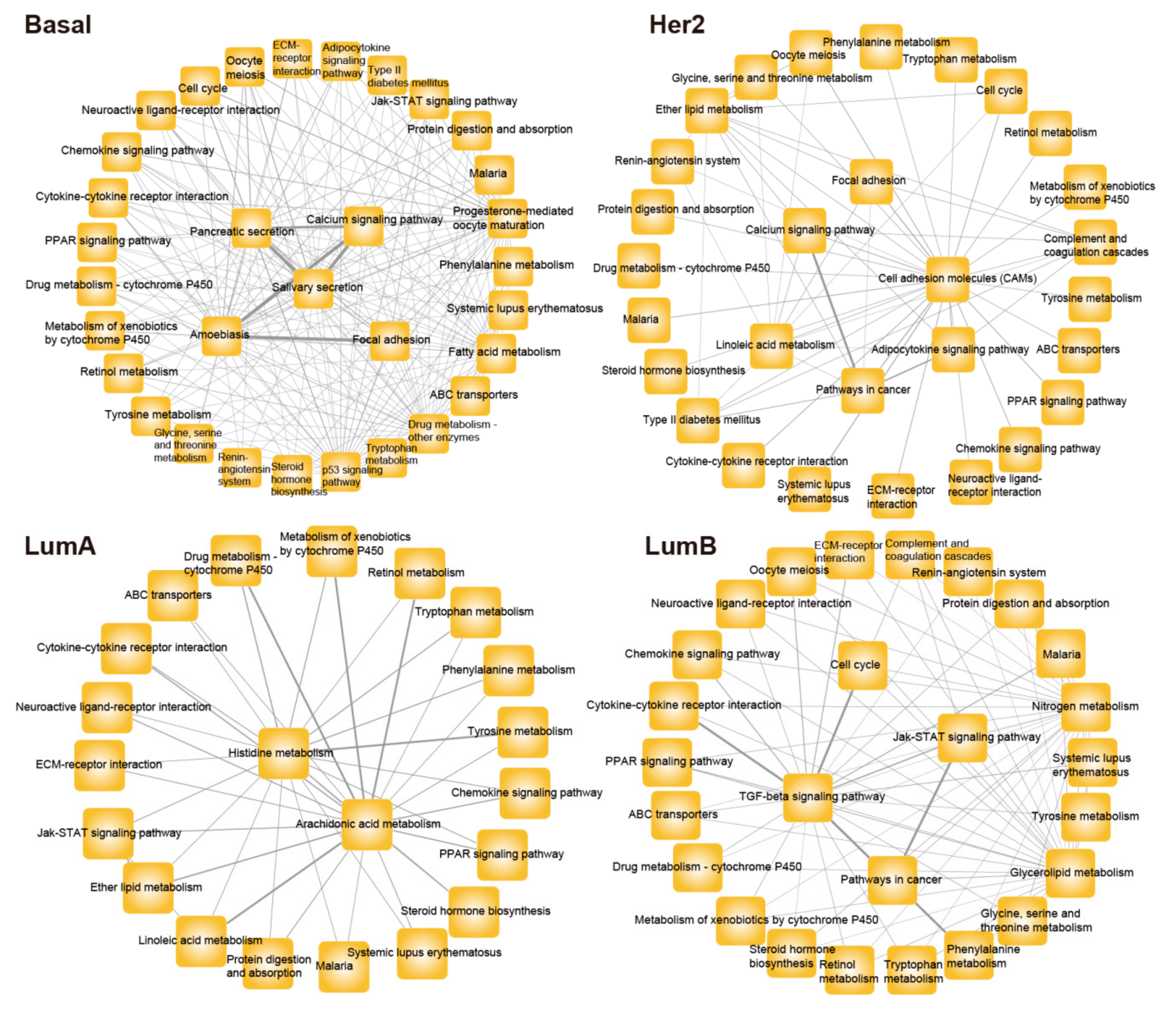
3.2. Constructing Risk Pathway Crosstalk Networks for Various Subtypes of Breast Cancer
3.3. Screening Candidate Therapeutic Drugs for Each Subtype of Breast Cancer Based on DS Score
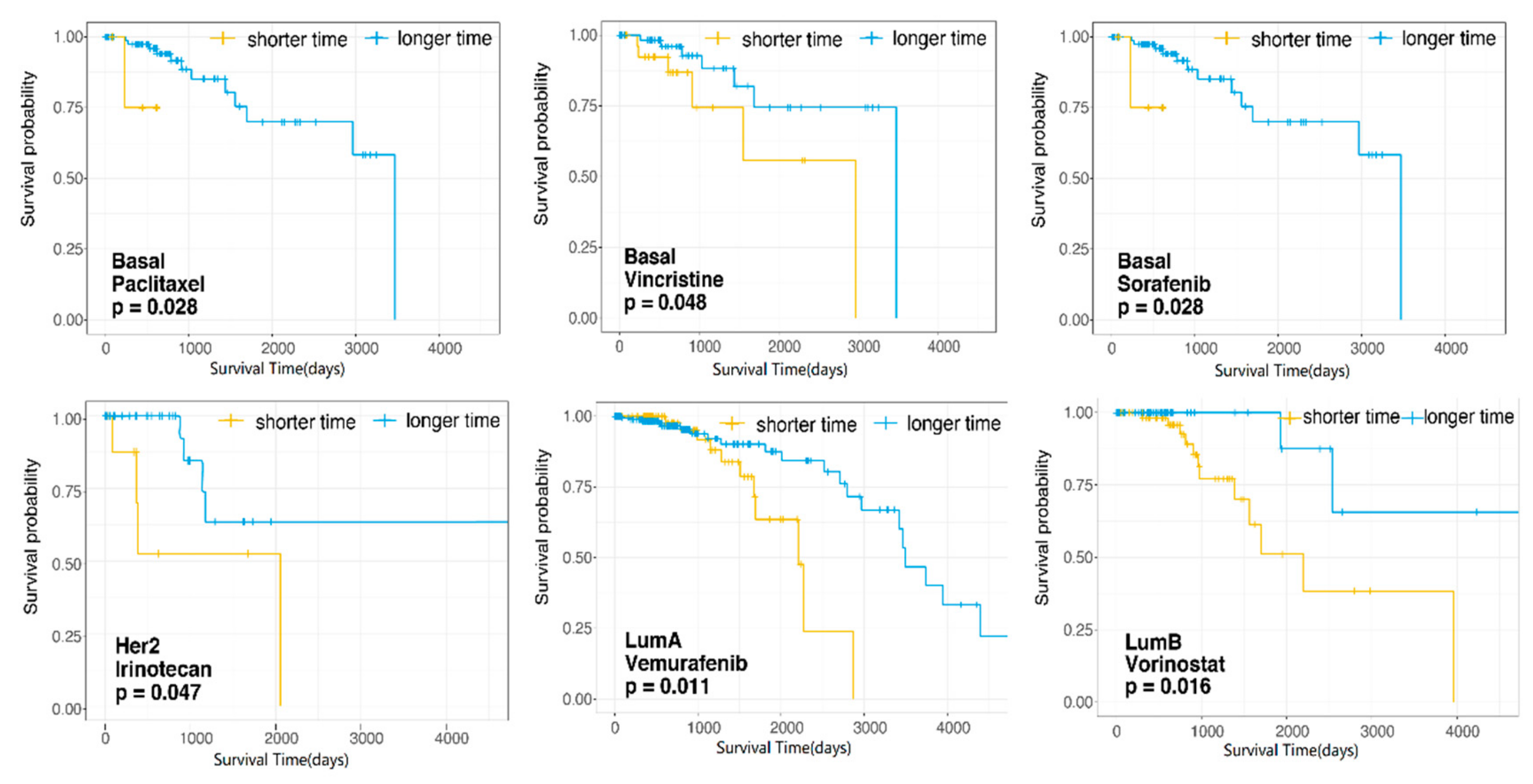
3.4. Dissecting the Effects of Candidate Therapeutic Drugs for Patient Survival in Each Subtype of Breast Cancer
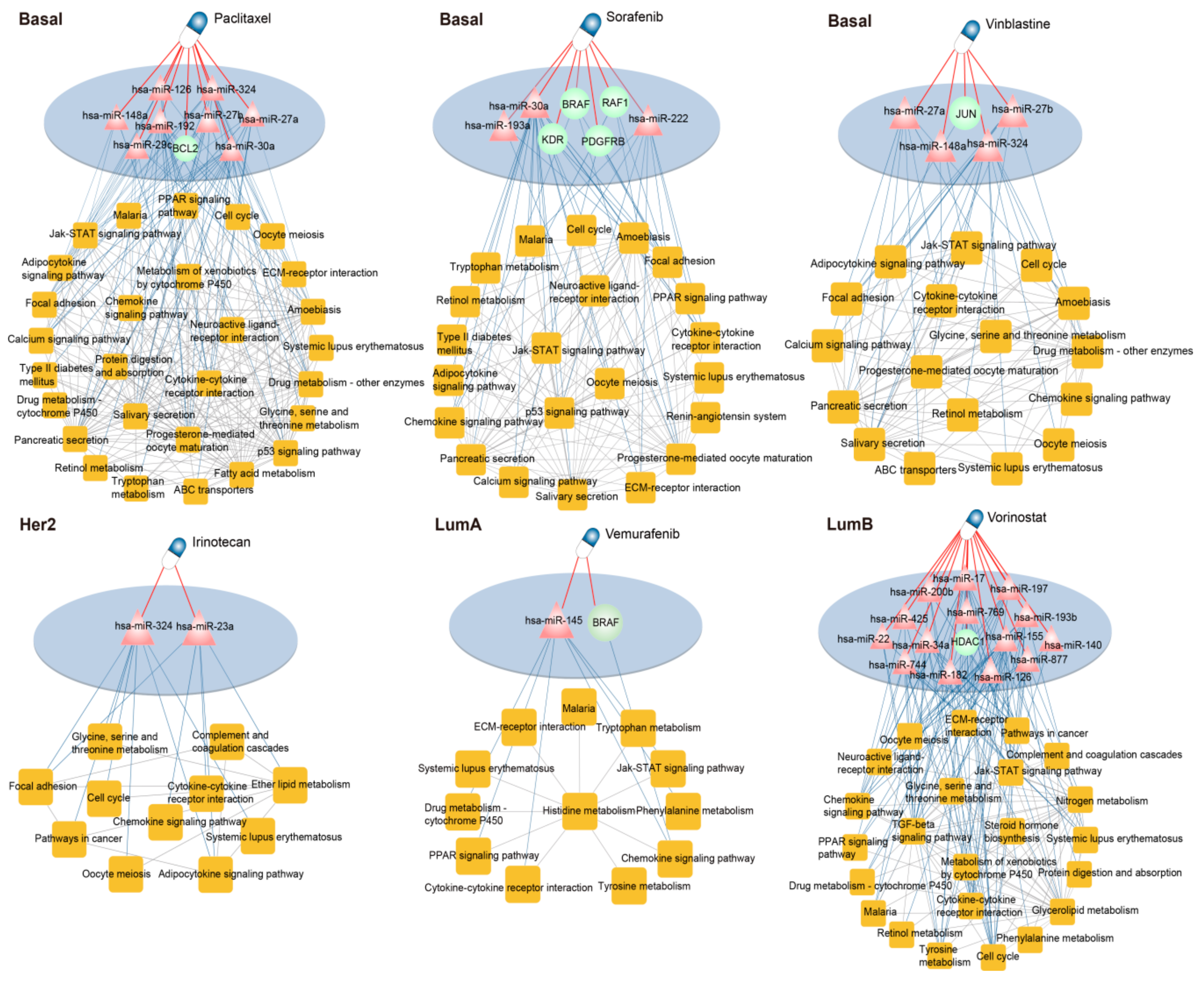
3.5. Dissecting the Mechanism of Candidate Drugs for Each Subtype
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Holm, J.; Eriksson, L.; Ploner, A.; Eriksson, M.; Rantalainen, M.; Li, J.; Hall, P.; Czene, K. Assessment of breast cancer risk factors reveals subtype heterogeneity. Cancer Res. 2017, 77, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J.; Panel Members. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the st. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on er/pr and her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.S. Recent advances in breast cancer treatment. Hong Kong Med. J. Xianggang Yi Xue Za Zhi 2018, 24, 6–8. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Zhao, H.; Hu, J.; Ping, Y.; Li, F.; Lan, Y.; Xu, C.; Xiao, Y.; Li, X. Identifying the crosstalk of dysfunctional pathways mediated by lncRNAs in breast cancer subtypes. Mol. BioSyst. 2016, 12, 711–720. [Google Scholar] [CrossRef]
- Restelli, M.; Magni, M.; Ruscica, V.; Pinciroli, P.; De Cecco, L.; Buscemi, G.; Delia, D.; Zannini, L. A novel crosstalk between ccar2 and akt pathway in the regulation of cancer cell proliferation. Cell Death Dis. 2016, 7, e2453. [Google Scholar] [CrossRef]
- Brechbiel, J.M.-M.K.; Adjei, A.A. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat. Rev. 2014, 40, 750–759. [Google Scholar] [CrossRef]
- Aksamitiene, E.; Kiyatkin, A.; Kholodenko, B.N. Cross-talk between mitogenic ras/mapk and survival pi3k/akt pathways: A fine balance. Biochem. Soc. Focus. Meet. 2012, 40, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Igea, A.; Arroyo, R.; Alcalde, V.; Canovas, B.; Orozco, M.; Nebreda, A.R.; Aloy, P. Quantification of pathway cross-talk reveals novel synergistic drug combinations for breast cancer. Cancer Res. 2017, 77, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Godard, P.; van Eyll, J. Pathway analysis from lists of microRNAs: Common pitfalls and alternative strategy. Nucleic Acids Res. 2015, 43, 3490–3497. [Google Scholar] [CrossRef] [PubMed]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef] [PubMed]
- Anton, R.; Chatterjee, S.S.; Simundza, J.; Cowin, P.; Dasgupta, R. A systematic screen for micro-RNAs regulating the canonical wnt pathway. PLoS ONE 2011, 6, e26257. [Google Scholar] [CrossRef] [PubMed]
- Kaboli, P.J.; Rahmat, A.; Ismail, P.; Ling, K.H. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol. Res. 2015, 97, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Yu, Z.; Gao, X.; Gong, J.; Fan, L.; Liu, F. Simvastatin induces breast cancer cell death through oxidative stress up-regulating mir-140-5p. Aging (Albany NY) 2019, 11, 3198–3219. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Li, Y.; Qi, X.; Guo, Y. Triiodothyronine promotes cell proliferation of breast cancer via modulating mir-204/amphiregulin. Pathol. Oncol. Res. POR 2019, 25, 653–658. [Google Scholar] [CrossRef]
- Shenoda, B.B.; Ramanathan, S.; Ajit, S.K. In vitro validation of miRNA-mediated gene expression linked to drug metabolism. Curr. Protoc. Pharmacol. 2017, 79, 9.26.1–9.26.15. [Google Scholar]
- Liu, X.; Wang, S.; Meng, F.; Wang, J.; Zhang, Y.; Dai, E.; Yu, X.; Li, X.; Jiang, W. Sm2mir: A database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics 2013, 29, 409–411. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Li, F.; Li, X.; Feng, L.; Shi, X.; Wang, L.; Li, X. Dissecting dysfunctional crosstalk pathways regulated by miRNAs during glioma progression. Oncotarget 2016, 7, 25769–25782. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.D.; Lin, F.M.; Wu, W.Y.; Liang, C.; Huang, W.C.; Chan, W.L.; Tsai, W.T.; Chen, G.Z.; Lee, C.J.; Chiu, C.M.; et al. Mirtarbase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. Mir2disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. Mirecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, D105–D110. [Google Scholar] [CrossRef] [PubMed]
- Vergoulis, T.; Vlachos, I.S.; Alexiou, P.; Georgakilas, G.; Maragkakis, M.; Reczko, M.; Gerangelos, S.; Koziris, N.; Dalamagas, T.; Hatzigeorgiou, A.G. Tarbase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012, 40, D222–D229. [Google Scholar] [CrossRef] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in kegg. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. Drugbank 5.0: A major update to the drugbank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Miao, Y.; Wang, Q.; Jiang, W.; Xu, C.; Li, J.; Han, J.; Zhang, F.; Gong, B.; et al. Subpathwayminer: A software package for flexible identification of pathways. Nucleic Acids Res. 2009, 37, e131. [Google Scholar] [CrossRef] [PubMed]
- Bernards, R. A missing link in genotype-directed cancer therapy. Cell 2012, 151, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Chang, S.S.; Hsu, J.L.; Hung, M.C. Signaling cross-talk in the resistance to her family receptor targeted therapy. Oncogene 2014, 33, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Koscielny, G.; An, P.; Carvalho-Silva, D.; Cham, J.A.; Fumis, L.; Gasparyan, R.; Hasan, S.; Karamanis, N.; Maguire, M.; Papa, E.; et al. Open targets: A platform for therapeutic target identification and validation. Nucleic Acids Res. 2017, 45, D985–D994. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Ju, R.J.; Liu, L.; Xie, H.J.; Mu, L.M.; Zhao, Y.; Yan, Y.; Hu, Y.J.; Wu, J.S.; Lu, W.L. Application of functional vincristine plus dasatinib liposomes to deletion of vasculogenic mimicry channels in triple-negative breast cancer. Oncotarget 2015, 6, 36625–36642. [Google Scholar] [CrossRef] [PubMed]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [PubMed]
- Wilhelm, S.; Carter, C.; Lynch, M.; Lowinger, T.; Dumas, J.; Smith, R.A.; Schwartz, B.; Simantov, R.; Kelley, S. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 2006, 5, 835–844. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, X.; Huo, Q.; Sun, M.; Cai, C.; Liu, Z.; Hu, G.; Yang, Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 2014, 33, 3119–3128. [Google Scholar] [CrossRef]
- Shah, M.Y.; Calin, G.A. MicroRNAs mir-221 and mir-222: A new level of regulation in aggressive breast cancer. Genome Med. 2011, 3, 56. [Google Scholar] [CrossRef]
- Tsai, K.W.; Leung, C.M.; Lo, Y.H.; Chen, T.W.; Chan, W.C.; Yu, S.Y.; Tu, Y.T.; Lam, H.C.; Li, S.C.; Ger, L.P.; et al. Arm selection preference of microRNA-193a varies in breast cancer. Sci. Rep. 2016, 6, 28176. [Google Scholar] [CrossRef]
- Vanhoefer, U.; Harstrick, A.; Achterrath, W.; Cao, S.; Seeber, S.; Rustum, Y.M. Irinotecan in the treatment of colorectal cancer: Clinical overview. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2001, 19, 1501–1518. [Google Scholar] [CrossRef] [PubMed]
- Kuo, W.T.; Yu, S.Y.; Li, S.C.; Lam, H.C.; Chang, H.T.; Chen, W.S.; Yu, C.C. MicroRNA-324 in human cancer: Mir-324-5p and mir-324-3p have distinct biological functions in human cancer. Anticancer Res. 2016, 36, 5189–5196. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Shehata, H.H. Breast tissue–based microRNA panel highlights microRNA-23a and selected target genes as putative biomarkers for breast cancer. Transl. Res. 2015, 165, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Bollag, G.; Tsai, J.; Zhang, J.; Zhang, C.; Ibrahim, P.; Nolop, K.; Hirth, P. Vemurafenib: The first drug approved for braf-mutant cancer. Nat. Rev. Drug Discov. 2012, 11, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Mo, Y.Y. Mir-145-mediated suppression of cell growth, invasion and metastasis. Am. J. Transl. Res. 2010, 2, 170–180. [Google Scholar] [PubMed]
- Mann, B.S.; Johnson, J.R.; He, K.; Sridhara, R.; Abraham, S.; Booth, B.P.; Verbois, L.; Morse, D.E.; Jee, J.M.; Pope, S.; et al. Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous t-cell lymphoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 2318–2322. [Google Scholar] [CrossRef]
- Li, L.; Xie, X.; Luo, J.; Liu, M.; Xi, S.; Guo, J.; Tang, J. Targeted expression of mir-34a using the t-visa system suppresses breast cancer cell growth and invasion. Mol. Ther. 2012, 20, 2326–2334. [Google Scholar] [CrossRef]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.M.; Callen, D.F. The oncogenic role of mir-155 in breast cancer. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2012, 21, 1236–1243. [Google Scholar] [CrossRef]
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of aib1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201. [Google Scholar] [CrossRef]
- Chen, B.; Tang, H.; Liu, X.; Liu, P.; Yang, L.; Xie, X.; Wei, W. Mir-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015, 356, 410–417. [Google Scholar] [CrossRef]
- Li, Q.; Yao, Y.; Eades, G.; Liu, Z.; Zhang, Y.; Zhou, Q. Downregulation of mir-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene 2014, 33, 2589–2600. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Goding Sauer, A.; Newman, L.A.; Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Ayers, D.; Vandesompele, J. Influence of microRNAs and long non-coding RNAs in cancer chemoresistance. Genes 2017, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Zuzic, M.; Rojo Arias, J.E.; Wohl, S.G.; Busskamp, V. Retinal miRNA functions in health and disease. Genes 2019, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Reich, M. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, A.; Stein, G.Y.; Ruppin, E.; Sharan, R. PREDICT: A method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011, 7, 496. [Google Scholar] [CrossRef] [PubMed]
- Malas, T.B.; Vlietstra, W.J.; Kudrin, R.; Starikov, S.; Charrout, M.; Roos, M.; van Mulligen, E.M. Drug prioritization using the semantic properties of a knowledge graph. Sci. Rep. 2019, 9, 6281. [Google Scholar] [CrossRef] [PubMed]

| DS Score Ranking | Basal | Her2 | LumA | LumB |
|---|---|---|---|---|
| 1 | 5-Fluorouracil | Arsenic trioxide | Arsenic trioxide | Arsenic trioxide |
| 2 | Arsenic trioxide | Adriamycin | 5-Fluorouracil | Adriamycin |
| 3 | Tamoxifen | 5-Fluorouracil | Adriamycin | 5-Fluorouracil |
| 4 | Trastuzumab | Trastuzumab | Trastuzumab | Trastuzumab |
| 5 | Etoposide | Paclitaxel | Etoposide | Etoposide |
| 6 | Cisplatin | Temozolomide | Tamoxifen | Cisplatin |
| 7 | Paclitaxel | Etoposide | Vorinostat | Topotecan |
| 8 | Vorinostat | Gemcitabine | Bicalutamide | Irinotecan |
| 9 | Gemcitabine | Everolimus | Cisplatin | Paclitaxel |
| 10 | Adriamycin | Sunitinib | Vemurafenib | Tamoxifen |
| 11 | Temozolomide | Tamoxifen | Medroxyprogesterone acetate | Vemurafenib |
| 12 | Cyclophosphamide | Vorinostat | Gemcitabine | Gemcitabine |
| 13 | Bicalutamide | Cisplatin | Temozolomide | Sunitinib |
| 14 | Sunitinib | Sorafenib | Everolimus | Vorinostat |
| 15 | Vemurafenib | Cyclophosphamide | Sunitinib | Temozolomide |
| 16 | Medroxyprogesterone acetate | Goserelin | Paclitaxel | Everolimus |
| 17 | Everolimus | Vemurafenib | Oxaliplatin | Lenalidomide |
| 18 | Vinblastine | Bicalutamide | Cyclophosphamide | Cyclophosphamide |
| 19 | Lenalidomide | Vinblastine | Sorafenib | Bicalutamide |
| 20 | Oxaliplatin | Lenalidomide | Irinotecan | Goserelin |
| 21 | Sorafenib | Imatinib mesylate | Topotecan | Rapamycin |
| 22 | Goserelin | Bortezomib | Lenalidomide | Oxaliplatin |
| 23 | Irinotecan | Oxaliplatin | Vinblastine | |
| 24 | Mitoxantrone | Medroxyprogesterone acetate | Sorafenib | |
| 25 | Topotecan | Melphalan | Vincristine | |
| 26 | Imatinib mesylate | Gefitinib | Medroxyprogesterone acetate | |
| 27 | Vincristine | Rapamycin | Bortezomib | |
| 28 | Gefitinib | Vincristine | Imatinib mesylate | |
| 29 | Docetaxel | Irinotecan | Mitoxantrone | |
| 30 | Bortezomib | Topotecan | Melphalan | |
| 31 | Melphalan | Mitoxantrone | ||
| 32 | Rapamycin | Docetaxel | ||
| 33 | Epirubicin |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Lin, S.; Zhao, H.; Wang, J.; Zhang, C.; Dong, Q.; Hu, C.; Shang, D.; Wang, L.; Xu, Y. Quantifying Risk Pathway Crosstalk Mediated by miRNA to Screen Precision drugs for Breast Cancer Patients. Genes 2019, 10, 657. https://doi.org/10.3390/genes10090657
Xu Y, Lin S, Zhao H, Wang J, Zhang C, Dong Q, Hu C, Shang D, Wang L, Xu Y. Quantifying Risk Pathway Crosstalk Mediated by miRNA to Screen Precision drugs for Breast Cancer Patients. Genes. 2019; 10(9):657. https://doi.org/10.3390/genes10090657
Chicago/Turabian StyleXu, Yingqi, Shuting Lin, Hongying Zhao, Jingwen Wang, Chunlong Zhang, Qun Dong, Congxue Hu, Desi Shang, Li Wang, and Yanjun Xu. 2019. "Quantifying Risk Pathway Crosstalk Mediated by miRNA to Screen Precision drugs for Breast Cancer Patients" Genes 10, no. 9: 657. https://doi.org/10.3390/genes10090657
APA StyleXu, Y., Lin, S., Zhao, H., Wang, J., Zhang, C., Dong, Q., Hu, C., Shang, D., Wang, L., & Xu, Y. (2019). Quantifying Risk Pathway Crosstalk Mediated by miRNA to Screen Precision drugs for Breast Cancer Patients. Genes, 10(9), 657. https://doi.org/10.3390/genes10090657





