Genome-Wide Profiling of Enterotoxigenic Staphylococcus aureus Strains Used for the Production of Naturally Contaminated Cheeses
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Study Isolates and Sample Preparation
2.2. Detection of Staphylococcal Enterotoxin Genes by Multiplex PCR Assay, Typing and Antimicrobial Susceptibility Testing
2.3. Whole-Genome Sequencing, and Bioinformatics Analysis
2.4. Identification of Virulence Factors and Genomic Analysis
2.5. Production of the Natural Contaminated Cheese and Detection of Preformed Staphylococcal Enterotoxins in Milk and Cheese
3. Results
3.1. SEs Genes and Molecular Typing
3.2. Antimicrobial Susceptibility Testing
3.3. General Features of Enterotoxigenic S. aureus
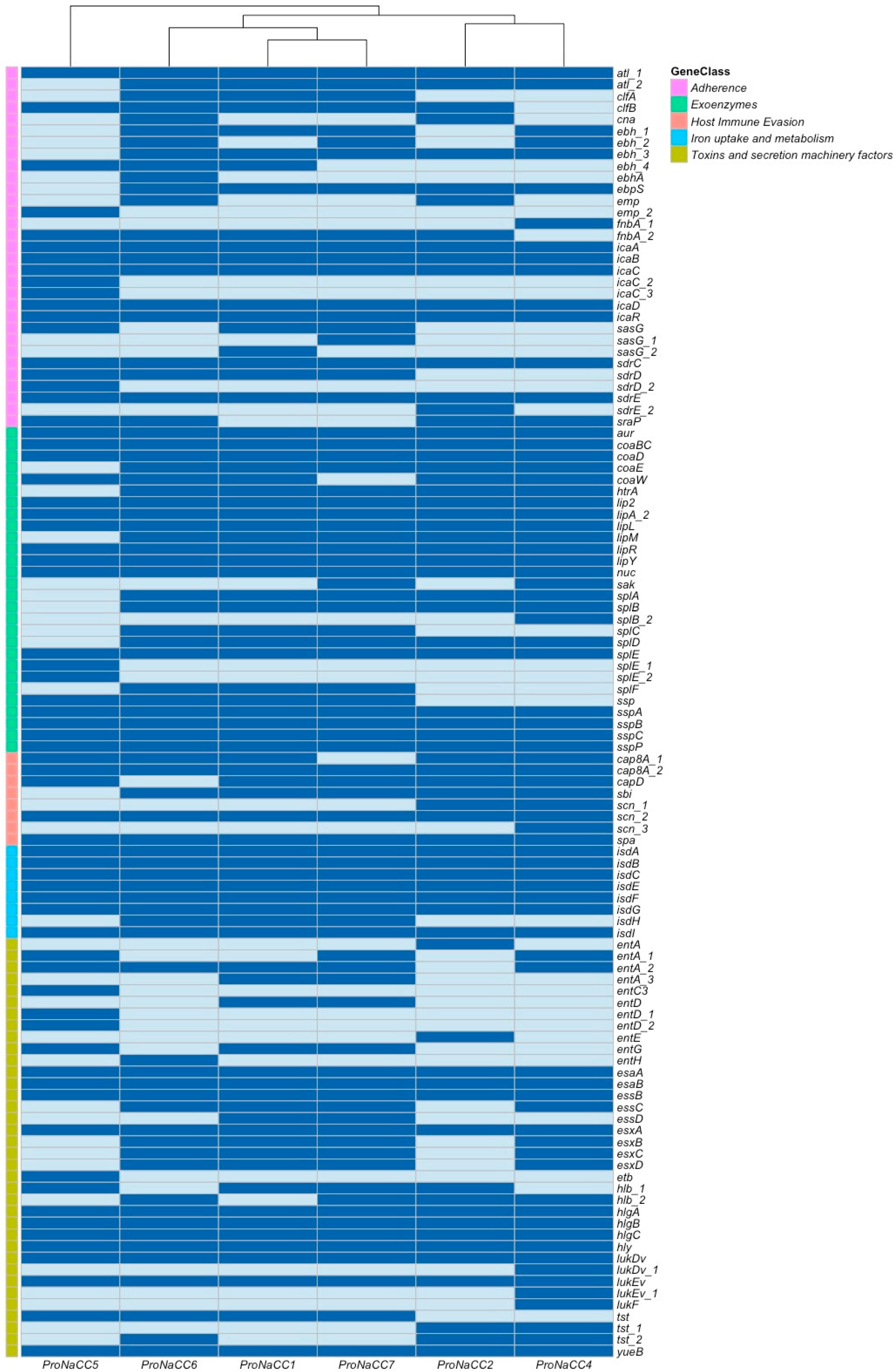
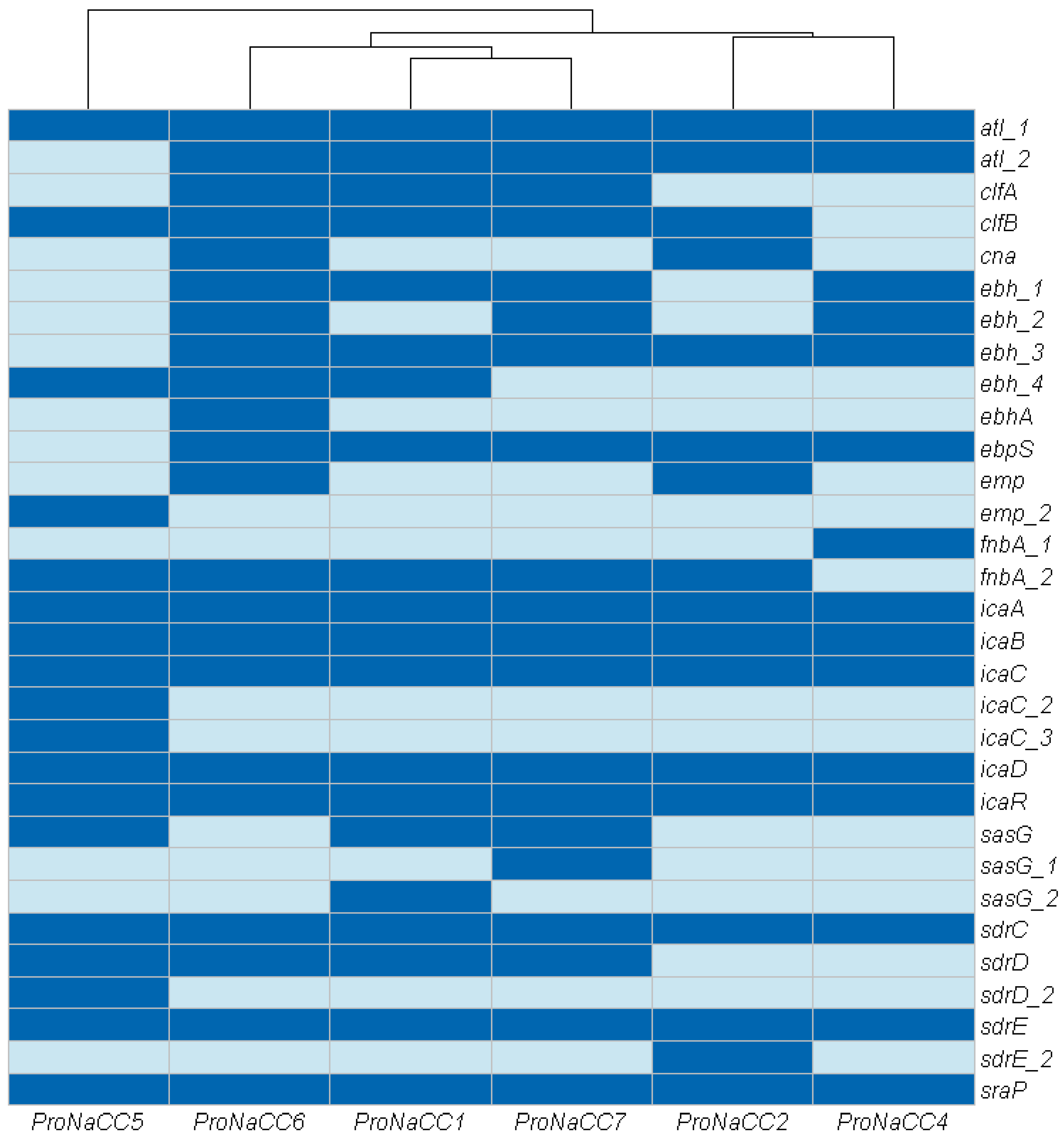
3.3.1. Adherence Factors
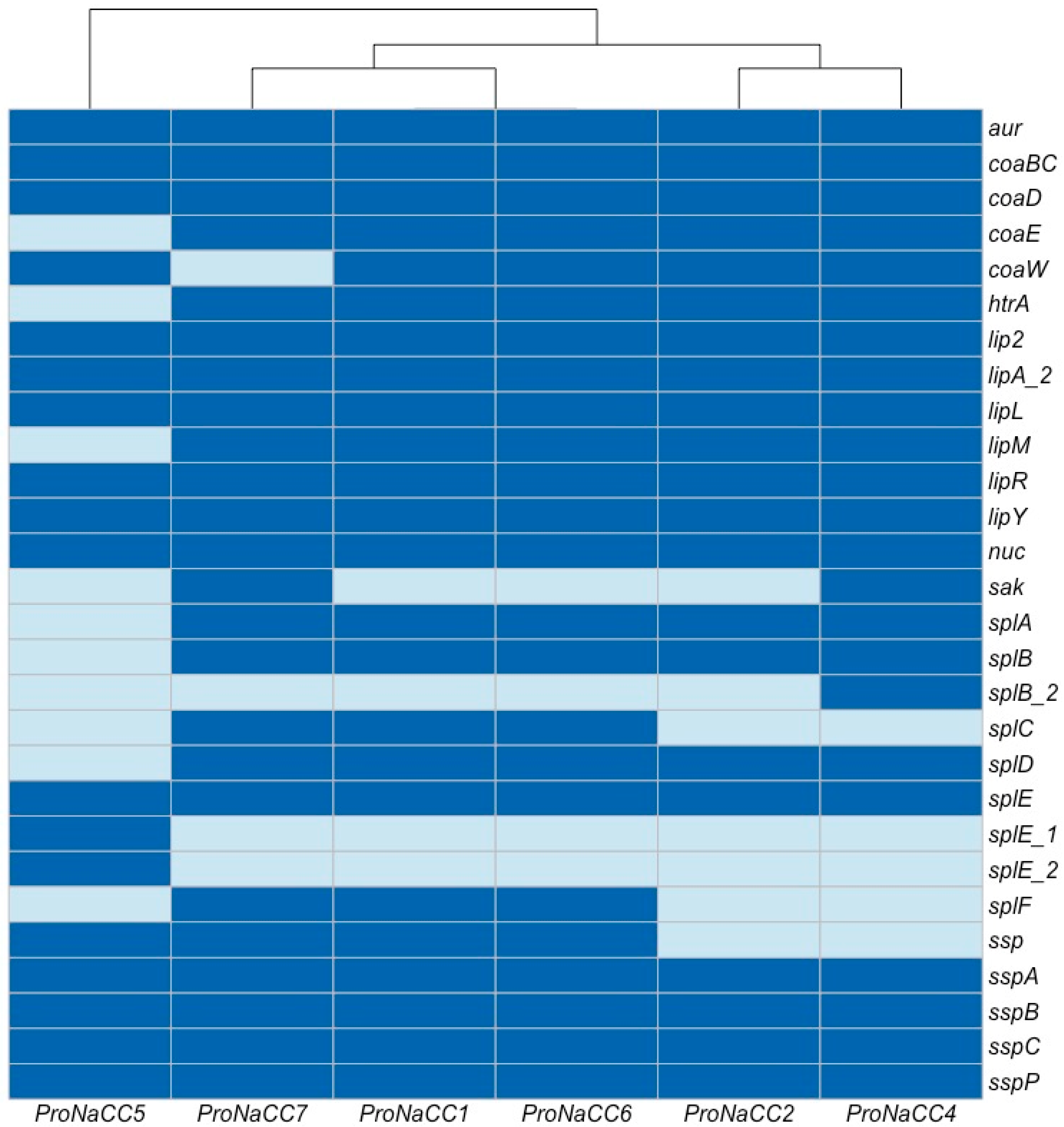
3.3.2. S. aureus Exoenzymes
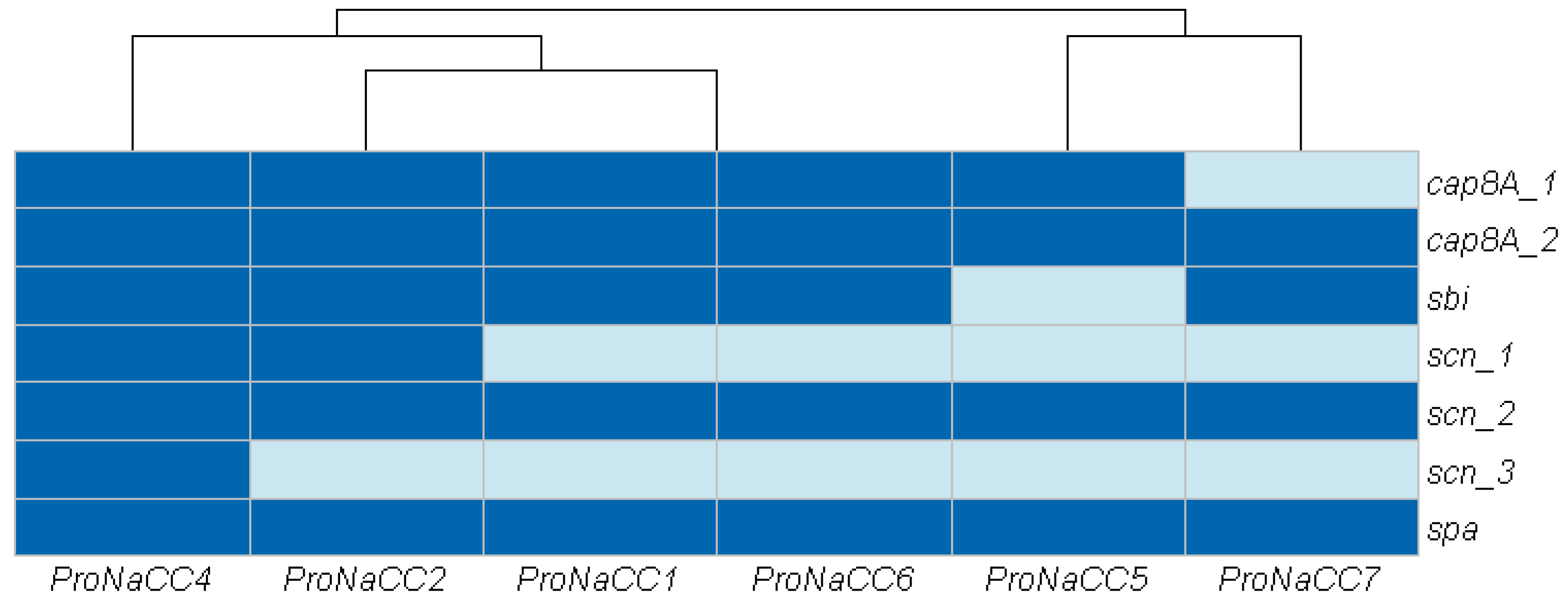
3.3.3. Genes Involved in Host Immune System Evasion
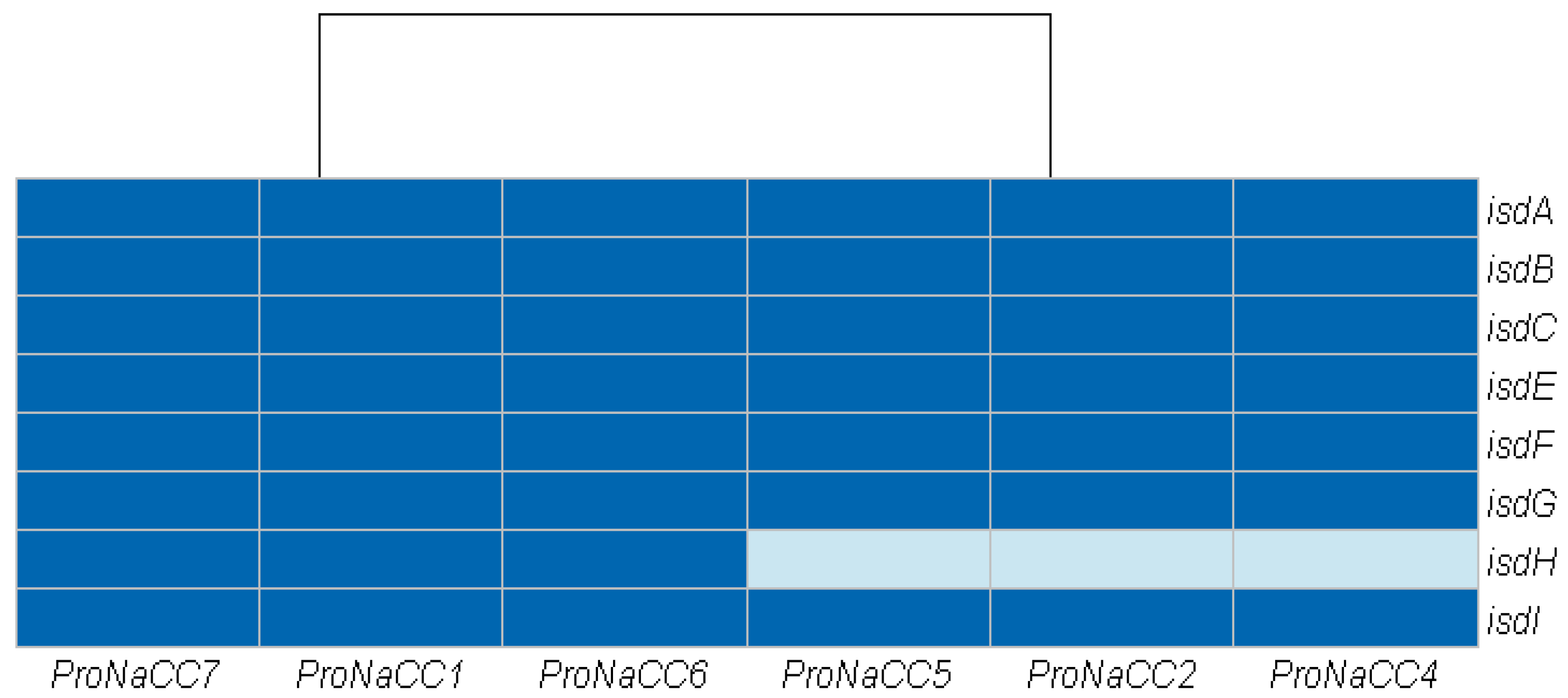
3.3.4. Iron Uptake Regulatory System and Metabolism
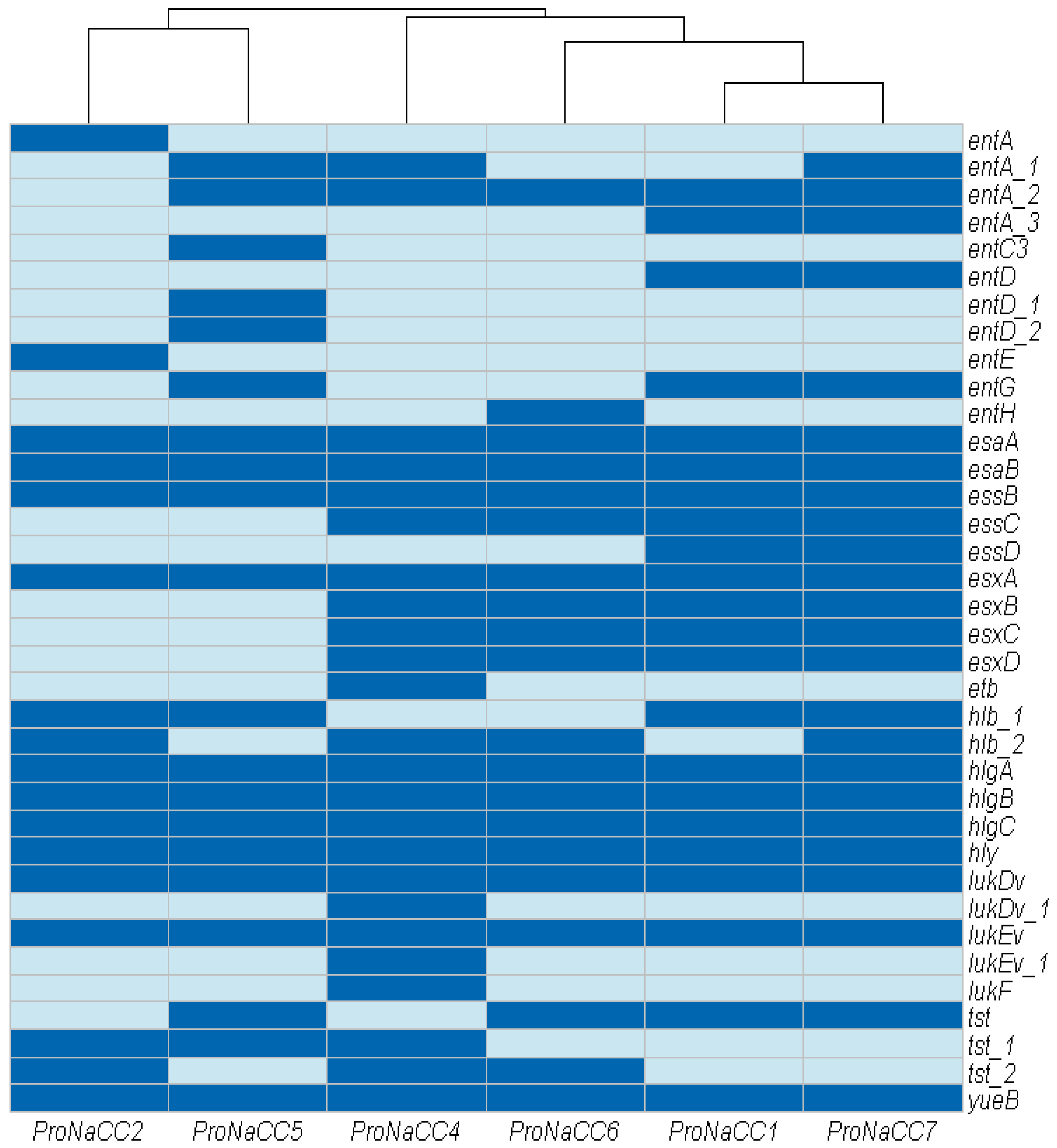
3.3.5. Toxins and Secretion Machinery Factors
3.3.6. Transcriptional Regulatory Elements
3.4. Enterotoxin Insight
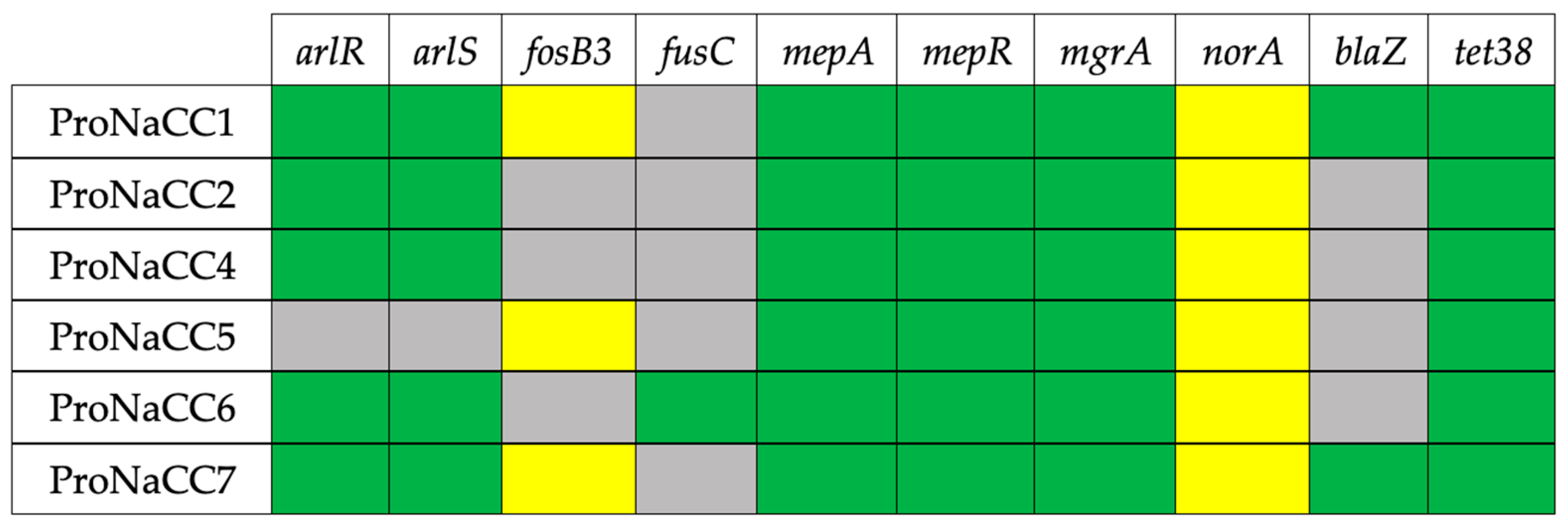
3.5. Antimicrobial Resistance Genes
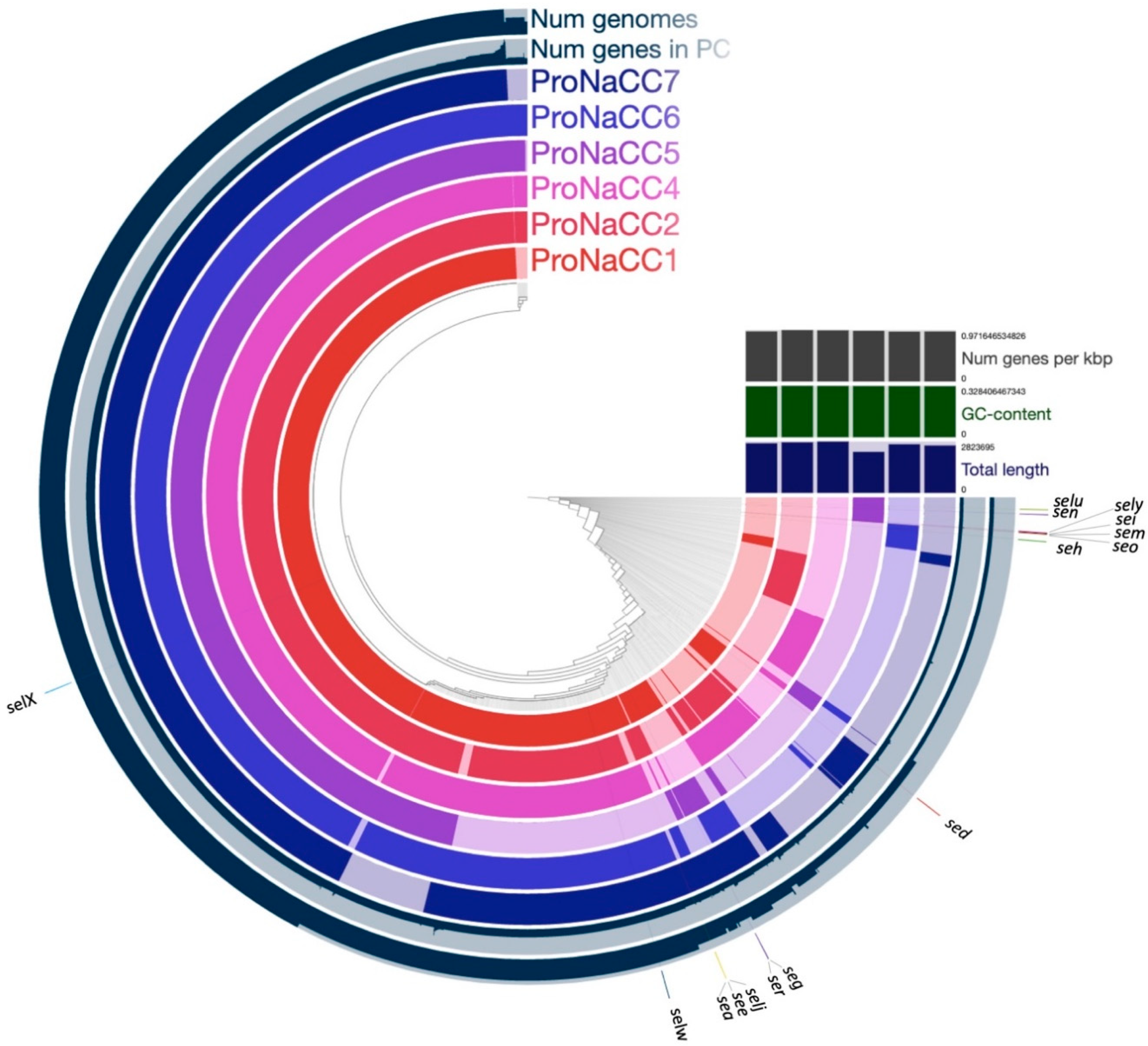
3.6. Core Genomes
3.7. SEs Production and Naturally-Contaminated Cheeses
4. Discussion
5. Data Access
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- MacLean, D.; Jones, J.D.; Studholme, D.J. Application of’next-generation’sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 2009, 7, 287. [Google Scholar] [CrossRef] [PubMed]
- Solieri, L.; Dakal, T.C.; Giudici, P. Next-generation sequencing and its potential impact on food microbial genomics. Ann. Microbiol. 2013, 63, 21–37. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Switt, A.I.M.; Wiedmann, M. Omics approaches in food safety: Fulfilling the promise? Trends Microbiol. 2014, 22, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef]
- Ward, T.J.; Ducey, T.F.; Usgaard, T.; Dunn, K.A.; Bielawski, J.P. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 2008, 74, 7629–7642. [Google Scholar] [CrossRef]
- Macori, G.; Cotter, P.D. Novel insights into the microbiology of fermented dairy foods. Curr. Opin. Biotechnol. 2017, 49, 172–178. [Google Scholar] [CrossRef]
- Button, J.E.; Dutton, R.J. Cheese microbes. Curr. Biol. 2012, 22, R587–R589. [Google Scholar] [CrossRef]
- Alemayehu, D.; Ross, R.P.; O’Sullivan, O.; Coffey, A.; Stanton, C.; Fitzgerald, G.F.; McAuliffe, O. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 2009, 448, 29–39. [Google Scholar] [CrossRef]
- Holden, M.T.; Hsu, L.Y.; Kurt, K.; Weinert, L.A.; Mather, A.E.; Harris, S.R.; Strommenger, B.; Layer, F.; Witte, W.; de Lencastre, H.; et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013, 23, 653–664. [Google Scholar] [CrossRef]
- Brunelle, B.W.; Bearson, B.L.; Bearson, S.M.D.; Casey, T.A. Multidrug-Resistant Salmonella enterica Serovar Typhimurium Isolates Are Resistant to Antibiotics That Influence Their Swimming and Swarming Motility. MSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, G.; Na, E.J.; Chung, H.Y.; Kim, S.; Kim, Y.T.; Kwak, W.; Kim, H.; Ryu, S.; Choi, S.H.; Lee, J.H. Genomic Insights and Its Comparative Analysis with Yersinia enterocolitica Reveals the Potential Virulence Determinants and Further Pathogenicity for Foodborne Outbreaks. J. Microbiol. Biotechnol. 2017, 27, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Köser, C.U.; Holden, M.T.; Ellington, M.J.; Cartwright, E.J.; Brown, N.M.; Ogilvy-Stuart, A.L.; Hsu, L.Y.; Chewapreecha, C.; Croucher, N.J.; Harris, S.R. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 2012, 366, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; He, Y.; Reed, S.; Strobaugh, T., Jr.; Irwin, P. Whole genome sequencing and analysis of Campylobacter coli YH502 from retail chicken reveals a plasmid-borne type VI secretion system. Genom. Data 2017, 11, 128–131. [Google Scholar] [CrossRef]
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 2017, 49, 224–229. [Google Scholar] [CrossRef]
- Li, Z.; Perez-Osorio, A.; Wang, Y.; Eckmann, K.; Glover, W.A.; Allard, M.W.; Brown, E.W.; Chen, Y. Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State. BMC Microbiol. 2017, 17, 134. [Google Scholar] [CrossRef]
- Nadon, C.; Van Walle, I.; Gerner-Smidt, P.; Campos, J.; Chinen, I.; Concepcion-Acevedo, J.; Gilpin, B.; Smith, A.M.; Man Kam, K.; Perez, E.; et al. PulseNet International: Vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef]
- Deng, X.; Shariat, N.; Driebe, E.M.; Roe, C.C.; Tolar, B.; Trees, E.; Keim, P.; Zhang, W.; Dudley, E.G.; Fields, P.I.; et al. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 2015, 53, 212–218. [Google Scholar] [CrossRef]
- Ransom, G.; Kaplan, B. USDA uses PulseNet for food safety. J. Am. Vet. Med. Assoc. 1998, 213, 1107. [Google Scholar]
- Bergholz, T.M.; den Bakker, H.C.; Katz, L.S.; Silk, B.J.; Jackson, K.A.; Kucerova, Z.; Joseph, L.A.; Turnsek, M.; Gladney, L.M.; Halpin, J.L. Determination of evolutionary relationships of outbreak-associated Listeria monocytogenes strains of serotypes 1/2a and 1/2b by whole-genome sequencing. Appl. Environ. Microbiol. 2016, 82, 928–938. [Google Scholar] [CrossRef][Green Version]
- Roetzer, A.; Diel, R.; Kohl, T.A.; Rückert, C.; Nübel, U.; Blom, J.; Wirth, T.; Jaenicke, S.; Schuback, S.; Rüsch-Gerdes, S. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: A longitudinal molecular epidemiological study. PLoS Med. 2013, 10, e1001387. [Google Scholar] [CrossRef] [PubMed]
- Oakeson, K.F.; Wagner, J.M.; Rohrwasser, A.; Atkinson-Dunn, R. Whole-Genome Sequencing and Bioinformatic Analysis of Isolates from Foodborne Illness Outbreaks of Campylobacter jejuni and Salmonella enterica. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Authority, T.E.F.S. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2017, 15, 1–231. [Google Scholar] [CrossRef]
- Argudin, M.A.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Johler, S.; Weder, D.; Bridy, C.; Huguenin, M.C.; Robert, L.; Hummerjohann, J.; Stephan, R. Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. J. Dairy Sci. 2015, 98, 2944–2948. [Google Scholar] [CrossRef]
- Gallina, S.; Bianchi, D.M.; Bellio, A.; Nogarol, C.; Macori, G.; Zaccaria, T.; Biorci, F.; Carraro, E.; Decastelli, L. Staphylococcal poisoning foodborne outbreak: Epidemiological investigation and strain genotyping. J. Food Prot. 2013, 76, 2093–2098. [Google Scholar] [CrossRef]
- Regulation (EC) No. 2073/2005 of the European Parliament and of the Council of 15 November 2005 on Microbiological Criteria for Foodstuffs. 2005. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32005R2073&from=EN (accessed on 27 September 2019).
- Sato’o, Y.; Omoe, K.; Naito, I.; Ono, H.K.; Nakane, A.; Sugai, M.; Yamagishi, N.; Hu, D.L. Molecular epidemiology and identification of a Staphylococcus aureus clone causing food poisoning outbreaks in Japan. J. Clin. Microbiol. 2014, 52, 2637–2640. [Google Scholar] [CrossRef]
- Fetsch, A.; Contzen, M.; Hartelt, K.; Kleiser, A.; Maassen, S.; Rau, J.; Kraushaar, B.; Layer, F.; Strommenger, B. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int. J. Food Microbiol. 2014, 187, 1–6. [Google Scholar] [CrossRef]
- Macori, G.; Bellio, A.; Bianchi, D.M.; Gallina, S.; Adriano, D.; Zuccon, F.; Chiesa, F.; Acutis, P.L.; Casalinuovo, F.; Decastelli, L. Molecular Typing of Staphylococcus Aureus Isolate Responsible for Staphylococcal Poisoning Incident in Homemade Food. Ital. J. Food Saf. 2016, 5, 5736. [Google Scholar] [CrossRef]
- Benkerroum, N. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: An overview. Crit. Rev. Food Sci. Nutr. 2018, 58, 1943–1970. [Google Scholar] [CrossRef]
- Johler, S.; Giannini, P.; Jermini, M.; Hummerjohann, J.; Baumgartner, A.; Stephan, R. Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins 2015, 7, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Kobayashi, M.; Matsushita, S.; Uehara, S.; Kato, R.; Yusuke, S.O.; Ono, H.K.; Sadamasu, K.; Kai, A.; Kamata, Y. Detection of the staphylococcal enterotoxin D-like gene from staphylococcal food poisoning isolates over the last two decades in Tokyo. J. Vet. Med. Sci. 2015, 77, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, A.; Tang, J.; Tang, C.; Chen, J.; Liu, J. Identification and measurement of staphylococcal enterotoxin-like protein I (SE ll) secretion from Staphylococcus aureus clinical isolate. J. Appl. Microbiol. 2016, 121, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Grumann, D.; Nübel, U.; Bröker, B.M. Staphylococcus aureus toxins–their functions and genetics. Infect. Genet. Evol. 2014, 21, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.L.; Ono, H.K.; Isayama, S.; Okada, R.; Okamura, M.; Lei, L.C.; Liu, Z.S.; Zhang, X.C.; Liu, M.Y.; Cui, J.C.; et al. Biological characteristics of staphylococcal enterotoxin Q and its potential risk for food poisoning. J. Appl. Microbiol. 2017, 122, 1672–1679. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.K.; Hirose, S.; Naito, I.; Sato’o, Y.; Asano, K.; Hu, D.L.; Omoe, K.; Nakane, A. The emetic activity of staphylococcal enterotoxins, SEK, SEL, SEM, SEN and SEO in a small emetic animal model, the house musk shrew. Microbiol. Immunol. 2017, 61, 12–16. [Google Scholar] [CrossRef]
- Omoe, K.; Hu, D.-L.; Ono, H.K.; Shimizu, S.; Takahashi-Omoe, H.; Nakane, A.; Uchiyama, T.; Shinagawa, K.; Imanishi, K.I. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect. Immun. 2013, 81, 3627–3631. [Google Scholar] [CrossRef]
- Ono, H.K.; Sato’o, Y.; Narita, K.; Naito, I.; Hirose, S.; Hisatsune, J.; Asano, K.; Hu, D.L.; Omoe, K.; Sugai, M.; et al. Identification and Characterization of a Novel Staphylococcal Emetic Toxin. Appl. Environ. Microbiol. 2015, 81, 7034–7040. [Google Scholar] [CrossRef]
- Dearborn, A.D.; Wall, E.A.; Kizziah, J.L.; Klenow, L.; Parker, L.K.; Manning, K.A.; Spilman, M.S.; Spear, J.M.; Christie, G.E.; Dokland, T. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. Elife 2017, 6, e30822. [Google Scholar] [CrossRef]
- Wilson, G.J.; Tuffs, S.W.; Wee, B.A.; Seo, K.S.; Park, N.; Connelley, T.; Guinane, C.M.; Morrison, W.I.; Fitzgerald, J.R. Bovine Staphylococcus aureus superantigens stimulate the entire T cell repertoire of cattle. Infect. Immun. 2018, 86, e00505–e00518. [Google Scholar] [CrossRef]
- Thomas, D.; Dauwalder, O.; Brun, V.; Badiou, C.; Ferry, T.; Etienne, J.; Vandenesch, F.; Lina, G. Staphylococcus aureus superantigens elicit redundant and extensive human Vbeta patterns. Infect. Immun. 2009, 77, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Zapotoczna, M.; Riboldi, G.P.; Moustafa, A.M.; Dickson, E.; Narechania, A.; Morrissey, J.A.; Planet, P.J.; Holden, M.T.; Waldron, K.J.; Geoghegan, J.A. Mobile-Genetic-Element-Encoded Hypertolerance to Copper Protects Staphylococcus aureus from Killing by Host Phagocytes. MBio 2018, 9, e00550-18. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Alibayov, B.; Zdenkova, K.; Sykorova, H.; Demnerova, K. Molecular analysis of Staphylococcus aureus pathogenicity islands (SaPI) and their superantigens combination of food samples. J. Microbiol. Methods 2014, 107, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Haaber, J.; Penadés, J.R.; Ingmer, H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017, 25, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Zeleny, R.; Nia, Y.; Schimmel, H.; Mutel, I.; Hennekinne, J.A.; Emteborg, H.; Charoud-Got, J.; Auvray, F. Certified reference materials for testing of the presence/absence of Staphylococcus aureus enterotoxin A (SEA) in cheese. Anal. Bioanal. Chem. 2016, 408, 5457–5465. [Google Scholar] [CrossRef]
- Ostyn, A.; De Buyser, M.; Guillier, F.; Groult, J.; Felix, B.; Salah, S.; Delmas, G.; Hennekinne, J. First evidence of a food poisoning outbreak due to staphylococcal enterotoxin type E, France, 2009. Eurosurveillance 2010, 15, 19528. [Google Scholar]
- Bellio, A.; Chiesa, F.; Gallina, S.; Bianchi, D.M.; Macori, G.; Bossi, D.; Nia, Y.; Mutel, I.; Messio, S.; Hennekinne, J.A.; et al. Insight into the Distribution of Staphylococci and Their Enterotoxins in Cheeses Under Natural Conditions. Front. Microbiol. 2018, 9, 3233. [Google Scholar] [CrossRef]
- De Buyser, M.-L.; Grout, J.; Brisabois, A.; Assere, A.; Lombard, B. Detection of genes encoding staphylococcal enterotoxins. Mult. PCR Sea See Ser Method CRL Coagulase Posit. Staphylococci Incl. Staphylococcus Aureus 2009, 1–5. [Google Scholar]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones ofStaphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [PubMed]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC. Qual. Control Tool High Throughput Seq. Data 2010, 370. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 27 December 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2016, 45, D535–D542. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Montalban-Lopez, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief. Bioinform. 2017, 20. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2017, 34. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Ostyn, A.; Prufer, A.; Papinaud, I.; Hennekinne, J.; Assere, A.; Lombard, B. Detection of Staphylococcal Enterotoxins Types SEA to SEE in All Types of Food Matrices. In Proceedings of the European Screening Method of the EU–RL for Coagulase Positive Staphylococci including Staphylococcus, Aureus, France, September 2010; Version 5. pp. 1–12. [Google Scholar]
- International Organization for Standardization. ISO 6888-2 Microbiology of Food Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus Aureus and Other Species)—Part-2: Technique Using Rabbit Plasma Fibrynogen Agar Medium; International Organization for Standardization: Geneva, Switzerland, 1999; p. 7. [Google Scholar]
- Ohr, R.J.; Anderson, M.; Shi, M.; Schneewind, O.; Missiakas, D. EssD, a Nuclease Effector of the Staphylococcus aureus ESS Pathway. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Zoltner, M.; Ng, W.M.; Money, J.J.; Fyfe, P.K.; Kneuper, H.; Palmer, T.; Hunter, W.N. EssC: Domain structures inform on the elusive translocation channel in the Type VII secretion system. Biochem. J. 2016, 473, 1941–1952. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; Deboy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed]
- Abaev, I.; Skryabin, Y.; Kislichkina, A.; Bogun, A.; Korobova, O.; Mayskaya, N.; Shemyakin, I.; Dyatlov, I. Draft Genome Sequences of Exfoliative Toxin A-Producing Staphylococcus aureus Strains B-7772 and B-7777 (CC8/ST2993) and B-7774 (CC15/ST2126), Isolated in a Maternity Hospital in the Central Federal District of Russia. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Truong-Bolduc, Q.C.; Zhang, X.; Hooper, D.C. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 2003, 185, 3127–3138. [Google Scholar] [CrossRef] [PubMed]
- Daly, K.M.; Upton, M.; Sandiford, S.K.; Draper, L.A.; Wescombe, P.A.; Jack, R.W.; O’Connor, P.M.; Rossney, A.; Götz, F.; Hill, C.; et al. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J. Bacteriol. 2010, 192, 1131–1142. [Google Scholar] [CrossRef]
- Kérouanton, A.; Hennekinne, J.A.; Letertre, C.; Petit, L.; Chesneau, O.; Brisabois, A.; De Buyser, M.L. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 2007, 115, 369–375. [Google Scholar] [CrossRef]
- Guidi, F.; Duranti, A.; Gallina, S.; Nia, Y.; Petruzzelli, A.; Romano, A.; Travaglini, V.; Olivastri, A.; Calvaresi, V.; Decastelli, L.; et al. Characterization of A Staphylococcal Food Poisoning Outbreak in A Workplace Canteen during the Post-Earthquake Reconstruction of Central Italy. Toxins 2018, 10, 523. [Google Scholar] [CrossRef]
- Li, G.; Wu, S.; Luo, W.; Su, Y.; Luan, Y.; Wang, X. Staphylococcus aureus ST6-t701 isolates from food-poisoning outbreaks (2006–2013) in Xi’an, China. Foodborne Pathog. Dis. 2015, 12, 203–206. [Google Scholar] [CrossRef]
- Johler, S.; Tichaczek-Dischinger, P.S.; Rau, J.; Sihto, H.M.; Lehner, A.; Adam, M.; Stephan, R. Outbreak of Staphylococcal food poisoning due to SEA-producing Staphylococcus aureus. Foodborne Pathog. Dis. 2013, 10, 777–781. [Google Scholar] [CrossRef]
- Mossong, J.; Decruyenaere, F.; Moris, G.; Ragimbeau, C.; Olinger, C.M.; Johler, S.; Perrin, M.; Hau, P.; Weicherding, P. Investigation of a staphylococcal food poisoning outbreak combining case-control, traditional typing and whole genome sequencing methods, Luxembourg, June 2014. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef]
- Crovadore, J.; Calmin, G.; Tonacini, J.; Chablais, R.; Baumgartner, A.; Schnyder, B.; Hodille, E.; Lefort, F. Whole-Genome Sequences of 15 Strains of Staphylococcus aureus subsp. aureus Isolated from Foodstuff and Human Clinical Samples. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Naushad, S.; Naqvi, S.A.; Nobrega, D.; Luby, C.; Kastelic, J.P.; Barkema, H.W.; De Buck, J. Comprehensive Virulence Gene Profiling of Bovine Non-aureus Staphylococci Based on Whole-Genome Sequencing Data. MSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Bien, J.; Sokolova, O.; Bozko, P. Characterization of Virulence Factors of Staphylococcus aureus: Novel Function of Known Virulence Factors That Are Implicated in Activation of Airway Epithelial Proinflammatory Response. J. Pathog. 2011, 2011, 601905. [Google Scholar] [CrossRef] [PubMed]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The intercellular adhesion (ICA) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [PubMed]
- Rode, T.M.; Langsrud, S.; Holck, A.; Møretrø, T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. Int. J. Food Microbiol. 2007, 116, 372–383. [Google Scholar] [CrossRef]
- Møretrø, T.; Hermansen, L.; Holck, A.L.; Sidhu, M.S.; Rudi, K.; Langsrud, S. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 2003, 69, 5648–5655. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Miajlovic, H.; Foster, T.J. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009, 9, 22. [Google Scholar] [CrossRef]
- Sharp, J.A.; Echague, C.G.; Hair, P.S.; Ward, M.D.; Nyalwidhe, J.O.; Geoghegan, J.A.; Foster, T.J.; Cunnion, K.M. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE 2012, 7, e38407. [Google Scholar] [CrossRef]
- Åvall-Jääskeläinen, S.; Taponen, S.; Kant, R.; Paulin, L.; Blom, J.; Palva, A.; Koort, J. Comparative genome analysis of 24 bovine-associated. PeerJ 2018, 6, e4560. [Google Scholar] [CrossRef]
- Rigoulay, C.; Entenza, J.M.; Halpern, D.; Widmer, E.; Moreillon, P.; Poquet, I.; Gruss, A. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect. Immun. 2005, 73, 563–572. [Google Scholar] [CrossRef]
- Reed, S.B.; Wesson, C.A.; Liou, L.E.; Trumble, W.R.; Schlievert, P.M.; Bohach, G.A.; Bayles, K.W. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 2001, 69, 1521–1527. [Google Scholar] [CrossRef]
- Waryah, C.B.; Gogoi-Tiwari, J.; Wells, K.; Eto, K.Y.; Masoumi, E.; Costantino, P.; Kotiw, M.; Mukkur, T. Diversity of Virulence Factors Associated with West Australian Methicillin-Sensitive Staphylococcus aureus Isolates of Human Origin. BioMed Res. Int. 2016, 2016, 8651918. [Google Scholar] [CrossRef] [PubMed]
- Ahmadrajabi, R.; Layegh-Khavidaki, S.; Kalantar-Neyestanaki, D.; Fasihi, Y. Molecular analysis of immune evasion cluster (IEC) genes and intercellular adhesion gene cluster (ICA) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J. Prev. Med. Hyg. 2017, 58, E308–E314. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Abdelnour, A.; Arvidson, S.; Bremell, T.; Ryden, C.; Tarkowski, A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 1993, 61, 3879–3885. [Google Scholar] [PubMed]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front. Microbiol. 2018, 9, 436. [Google Scholar] [CrossRef]
- Dubrac, S.; Msadek, T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 2004, 186, 1175–1181. [Google Scholar] [CrossRef]
- Wilson, G.J.; Seo, K.S.; Cartwright, R.A.; Connelley, T.; Chuang-Smith, O.N.; Merriman, J.A.; Guinane, C.M.; Park, J.Y.; Bohach, G.A.; Schlievert, P.M.; et al. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 2011, 7, e1002271. [Google Scholar] [CrossRef]
- Ono, H.K.; Omoe, K.; Imanishi, K.; Iwakabe, Y.; Hu, D.L.; Kato, H.; Saito, N.; Nakane, A.; Uchiyama, T.; Shinagawa, K. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 2008, 76, 4999–5005. [Google Scholar] [CrossRef]
- Zhang, S.; Iandolo, J.J.; Stewart, G.C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 1998, 168, 227–233. [Google Scholar] [CrossRef]
- Lyon, B.R.; Skurray, R. Antimicrobial resistance of Staphylococcus aureus: Genetic basis. Microbiol. Rev. 1987, 51, 88–134. [Google Scholar] [PubMed]
- Roetzer, A.; Gruener, C.S.; Haller, G.; Beyerly, J.; Model, N.; Eibl, M.M. Enterotoxin Gene Cluster-Encoded SEI and SElN from Staphylococcus aureus Isolates are Crucial for the Induction of Human Blood Cell Proliferation and Pathogenicity in Rabbits. Toxins 2016, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Umeda, K.; Nakamura, H.; Yamamoto, K.; Nishina, N.; Yasufuku, K.; Hirai, Y.; Hirayama, T.; Goto, K.; Hase, A.; Ogasawara, J. Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int. J. Food Microbiol. 2017, 256, 30–35. [Google Scholar] [CrossRef] [PubMed]
- McLauchlin, J.; Narayanan, G.L.; Mithani, V.; O’Neill, G. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 2000, 63, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Omoe, K.; Ishikawa, M.; Shimoda, Y.; Hu, D.L.; Ueda, S.; Shinagawa, K. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates Harboring seg, seh, or sei genes. J. Clin. Microbiol. 2002, 40, 857–862. [Google Scholar] [CrossRef]
- Mira, A.; Martín-Cuadrado, A.B.; D’Auria, G.; Rodríguez-Valera, F. The bacterial pan-genome: A new paradigm in microbiology. Int. Microbiol. 2010, 13, 45–57. [Google Scholar] [CrossRef]
- Jamrozy, D.M.; Harris, S.R.; Mohamed, N.; Peacock, S.J.; Tan, C.Y.; Parkhill, J.; Anderson, A.S.; Holden, M.T.G. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb. Genom. 2016, 2, e000058. [Google Scholar] [CrossRef]
- Méric, G.; Miragaia, M.; de Been, M.; Yahara, K.; Pascoe, B.; Mageiros, L.; Mikhail, J.; Harris, L.G.; Wilkinson, T.S.; Rolo, J.; et al. Ecological Overlap and Horizontal Gene Transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol. 2015, 7, 1313–1328. [Google Scholar] [CrossRef]
- Roach, D.J.; Burton, J.N.; Lee, C.; Stackhouse, B.; Butler-Wu, S.M.; Cookson, B.T.; Shendure, J.; Salipante, S.J. A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PLoS Genet. 2015, 11, e1005413. [Google Scholar] [CrossRef]
- Bosi, E.; Monk, J.M.; Aziz, R.K.; Fondi, M.; Nizet, V.; Palsson, B. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl. Acad. Sci. USA 2016, 113, E3801–E3809. [Google Scholar] [CrossRef]
- Feil, E.J.; Cooper, J.E.; Grundmann, H.; Robinson, D.A.; Enright, M.C.; Berendt, T.; Peacock, S.J.; Smith, J.M.; Murphy, M.; Spratt, B.G.; et al. How clonal is Staphylococcus aureus? J. Bacterial. 2003, 185, 3307–3316. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.D.; Otto, M.; Braughton, K.R.; Whitney, A.R.; Chen, L.; Mathema, B.; Mediavilla, J.R.; Byrne, K.A.; Parkins, L.D.; Tenover, F.C.; et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc. Natl. Acad. Sci. USA 2008, 105, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Sihto, H.M.; Tasara, T.; Stephan, R.; Johler, S. Temporal expression of the staphylococcal enterotoxin D gene under NaCl stress conditions. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef]
- Sihto, H.M.; Stephan, R.; Engl, C.; Chen, J.; Johler, S. Effect of food-related stress conditions and loss of agr and sigB on seb promoter activity in S. aureus. Food Microbiol. 2017, 65, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.M.; Ingravalle, F.; Adriano, D.; Gallina, S.; Gramaglia, M.; Zuccon, F.; Astegiano, S.; Bellio, A.; Macori, G.; Ru, G.; et al. Reproducibility study for the detection of Staphylococcal enterotoxins in dairy products between official Italian national laboratories. J. Food Prot. 2014, 77, 999–1004. [Google Scholar] [CrossRef]
- Jørgensen, H.J.; Mathisen, T.; Løvseth, A.; Omoe, K.; Qvale, K.S.; Loncarevic, S. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol. Lett. 2005, 252, 267–272. [Google Scholar] [CrossRef]
- Pereira, M.L.; Carmo, L.S.D.; Santos, E.J.D.; Pereira, J.L.; Bergdoll, M.S. Enterotoxin H in Staphylococcal Food Poisoning. J. Food Prot. 1996, 59, 559–561. [Google Scholar] [CrossRef]



| Strain | Origin | Year | City, Country | Etiological Food | Patients | Symptoms 1 | Reference |
|---|---|---|---|---|---|---|---|
| ProNaCC1 | Routine analysis | 2009 | Haute-Savoie, France | Cheese (Tomme) | Not involved | Not reported | This study |
| ProNaCC2 | Staphylococcal Food-borne Poisoning | 2009 | Sommes, France | Cheese (Mont d’Or du Jura) | 23 | AC, D, F, N, V | [47] |
| ProNaCC4 | Staphylococcal Food-borne Poisoning | 2012 | Hautes-Vienne, France | Cheese (Basque Lait Cru Brebis) | 3 | AC, D, N, V | This study |
| ProNaCC5 | Staphylococcal Food-borne Poisoning | 2014 | Loire, France | Cheese (Raclette) | 5 | AC, D, V | This study |
| ProNaCC6 | Routine analysis | 2014 | Alpes de Haute Provence, France | Composed Salad (tuna-corn-beets-apple) | Not involved | Not reported | This study |
| ProNaCC7 | Routine analysis | 2015 | Vercelli, Italy | Cheese (Tomme) | Not involved | Not reported | [48] |
| Gene | Name Primer | Sequence (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| sea | GSEAR-1 | GGT TAT CAA TGT GCG GGT GG | 102 | [53] |
| GSEAR-2 | CGG CAC TTT TTT CTC TTC GG | |||
| seb | GSEBR-1 | GTA TGG TGG TGT AAC TGA GC | 164 | [53] |
| GSEBR-2 | CCA AAT AGT GAC GAG TTA GG | |||
| sec | GSECR-1 | AGA TGA AGT AGT TGA TGT GTA TGG | 451 | [53] |
| GSECR-2 | CAC ACT TTT AGA ATC AAC CG | |||
| sed | GSEDR-1 | CCA ATA ATA GGA GAA AAT AAA AG | 278 | [53] |
| GSEDR-2 | ATT GGT ATT TTT TTT CGT TC | |||
| see | SA-U | TGT ATG TAT GGA GGT GTA AC | 213 | [53] |
| SA-E rev | GCC AAA GCT GTC TGA G | |||
| ser | SER 1 | AGA TGT GTT TGG AAT ACC CTA T | 123 | [53] |
| SER 2 | CTA TCA GCT GTG GAG TGC AT | |||
| seg | SEG-F | GTT AGA GGA GGT TTT ATG | 198 | [54] |
| SEG-R | TTC CTT CAA CAG GTG GAG A | |||
| seh | SEH-F | CAA CTG CTG ATT TAG CTC AG | 173 | [54] |
| SEH-R | CCC AAA CAT TAG CAC CA | |||
| sei | SEI-F | GGC CAC TTT ATC AGG ACA | 328 | [54] |
| SEI-R | AAC TTA CAG GCA GTC CA | |||
| selj | SEJ-F | GTT CTG GTG GTA AAC CA | 131 | [54] |
| SEJ-R | GCG GAA CAA CAG TTC TGA | |||
| sep | SEP-F | TCA AAA GAC ACC GCC AA | 396 | [54] |
| SEP-R | ATT GTC CTT GAG CAC CA |
| Strain | Gene(s) | Spa-Type | MLST |
|---|---|---|---|
| S. aureus ProNaCC1 | sed | t2953 | ST-8 |
| S. aureus ProNaCC2 | see | t4461 | ST-425 |
| S. aureus ProNaCC4 | sea | t19075 1 | ST-581 |
| S. aureus ProNaCC5 | seg/sei | t164 | ST-389 |
| S. aureus ProNaCC6 | seh | t127 | ST-1 |
| S. aureus ProNaCC7 | sea/sed/selj/ser | t3802 | ST-8 |
| Strain | Antimicrobial | Class | MIC (mg/L) |
|---|---|---|---|
| ProNaCC1 | Benzylpenicillin | Penicillins | ≥0.5 |
| ProNaCC1 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| ProNaCC2 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| ProNaCC4 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| ProNaCC5 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| ProNaCC6 | Benzylpenicillin | Penicillins | ≥0.5 |
| ProNaCC6 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| ProNaCC7 | Enrofloxacin | Fluoroquinolones | ≤0.5 |
| Feature | ProNaCC1 | ProNaCC2 | ProNaCC4 | ProNaCC5 | ProNaCC6 | ProNaCC7 |
|---|---|---|---|---|---|---|
| Size (bp) | 2,728,931 | 2,782,905 | 2,823,695 | 2,260,169 | 2,677,370 | 2,607,693 |
| Contigs (>500 bp) | 19 | 45 | 54 | 15 | 9 | 14 |
| Contigs > 1 kb | 19 | 40 | 35 | 11 | 8 | 14 |
| CDs 1 | 2570 | 26,701 | 2747 | 2126 | 2473 | 2463 |
| G/C content | 32.53% | 32.80% | 32.78% | 32.71% | 32.66% | 32.61% |
| Strain | Enterotoxin | Contig | Sequence Start 1 | Sequence End 2 | % Coverage 3 | % Identity 4 | Reference 5 |
|---|---|---|---|---|---|---|---|
| ProNaCC1 | SED | 14 | 12,761 | 13,533 | 99.87 | 84.73 | UniProtKB—R9SA89 |
| SElJ | 14 | 14,428 | 15,212 | 97.64 | 83.95 | UniProtKB—O85217 | |
| SER | 14 | 15,329 | 16,103 | 99.74 | 84.9 | UniProtKB—Q76LS8 | |
| SElX | 3 | 249,399 | 250,007 | 100 | 84.24 | UniProtKB—G0Z026 | |
| SElW | 5 | 93,584 | 94,369 | 100 | 100 | GB—KX655710.1 | |
| ProNaCC2 | SEE | 1 | 409,445 | 410,214 | 99.87 | 83.9 | GB—WP_044122767 |
| SElX | 24 | 4093 | 4701 | 100 | 84.24 | UniProtKB—G0Z026 | |
| SElW | 6 | 46,777 | 47,558 | 99.36 | 96.55 | GB—KX655711.1| | |
| ProNaCC4 | SEA | 1 | 983 | 1744 | 98.57 | 81.15 | UniProtKB—P0A0L2 |
| SElW | 3 | 200,805 | 201,585 | 99.36 | 96.16 | GB—KX655711.1 | |
| SElX | 7 | 89,367 | 89,975 | 100 | 84.4 | UniProtKB—G0Z026 | |
| ProNaCC5 | SElY | 2 | 58,778 | 59,440 | 100 | 84.77 | UniProtKB—A0A0K2S2V0 |
| SElX | 2 | 634,655 | 635,263 | 100 | 84.56 | UniProtKB—G0Z025 | |
| SEG | 9 | 13,778 | 14,550 | 99.87 | 86.16 | UniProtKB—P0A0L8 | |
| SEN | 9 | 14,836 | 15,609 | 100 | 85.92 | UniProtKB—A0A0H3JS72 | |
| SElU | 9 | 15,630 | 16,397 | 98.08 | 84.42 | UniProtKB—Q6XXM3 | |
| SEI | 9 | 16,560 | 17,279 | 99.17 | 87.5 | UniProtKB—O85383 | |
| SEM | 9 | 17,320 | 18,033 | 99.58 | 85.01 | UniProtKB—A0A0H3K005 | |
| SEO | 9 | 18,317 | 19,096 | 100 | 87.82 | UniProtKB—A0A0H3JS76 | |
| ProNaCC6 | SElW | 1 | 1,068,149 | 1,068,933 | 99.87 | 97.84 | GB—KX655711.1 |
| SElX | 2 | 96,391 | 96,999 | 100 | 82.92 | UniProtKB—G0Z026 | |
| SEH | 5 | 98,979 | 99,700 | 99.86 | 86.29 | UniProtKB—P0A0M0 | |
| ProNaCC7 | SED | 11 | 6058 | 6830 | 99.87 | 84.73 | UniProtKB—R9SA89 |
| SElJ | 11 | 7725 | 8509 | 97.64 | 83.95 | UniProtKB—O85217 | |
| SER | 11 | 8626 | 9400 | 99.74 | −84.9 | UniProtKB—Q76LS8 | |
| SEA | 2 | 88,812 | 89,579 | 99.61 | 85.81 | UniProtKB—P0A0L2 | |
| SElW | 5 | 93,897 | 94,682 | 100 | 100 | GB—KX655710.1 | |
| SElX | 7 | 241,512 | 242,120 | 100 | 84.24 | UniProtKB—G0Z026 |
| Strain | SaPI1 1 | SaPI3 2 | SaPI3 3 | SaPI3 4 | SaPI3 5 | SaPI (fhuD) 6 | SaPIbov 7 | Prophage (Gene SE) | Plasmid pLUH02 8 | Plasmid pSK67 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| ProNaCC1 | n.d. | 100% | 100% | n.d. | n.d. | n.d. | n.d. | n.d. | 98.11% (sed, selj ser) | n.d. |
| ProNaCC2 | 1.46% | 100% | n.d. | 48.07% | 100% | n.d. | n.d. | ФN315, ФSa119 (see) | n.d. | n.d. |
| ProNaCC4 | n.d. | n.d. | n.d. | n.d. | 100% | 0.40% | 25.11% | ФN315, ФBU01 (sea) | n.d. | n.d. |
| ProNaCC5 | n.d. | 100% | n.d. | n.d. | n.d. | 23.98% | 0.44% | n.d. | n.d. | n.d. |
| ProNaCC6 | n.d. | 100% | 100% | n.d. | n.d. | n.d. | 0.44% | n.d. | n.d. | n.d. |
| ProNaCC7 | n.d. | 100% | 100% | n.d. | n.d. | n.d. | n.d. | ФN315, ФSa119, ФNM3, ФBU01 (sea) | n.d. | 85.14% (sed, selj ser) |
| Batch and Strain | SEs Genes Detected | Milk after 18 h Incubation | Sampling Area | Qualitative Methods | Amount of Protein Produced [SE] (ng/g of Cheese) a |
|---|---|---|---|---|---|
| 1—ProNaCC1 | sed/ser/selj | pos | Core | pos | [SED] = 7.966 |
| Periphery | pos | [SED] = 6.607 | |||
| 2—ProNaCC2 | see | pos | Core | pos | [SEE] = 9.126 |
| Periphery | pos | [SEE] = 8.419 | |||
| 3—ProNaCC4 | sea | pos | Core | pos | [SEA] = 2.760 |
| Periphery | pos | [SEA] = 2.648 | |||
| 4—ProNaCC5 | seg/sei | neg | Core | neg | neg |
| Periphery | neg | neg | |||
| 5—ProNaCC6 | seh | neg | Core | neg | neg |
| Periphery | neg | neg | |||
| 6—ProNaCC7 | sea/sed/selj/ser | pos | Core | pos | [SEA] = 1.833 |
| [SED] = 7.578 | |||||
| pos | Periphery | pos | [SEA] = 1.849 | ||
| [SED] = 7.841 |
| Batch and Strain | CPS in Spiked Milk 18 h Enrichment (CFU/g) | CPS in Fresh Cheese after Sweating (CFU/g) |
|---|---|---|
| 1—ProNaCC1 | 2.1 × 107 | 2.8 × 108 |
| 2—ProNaCC2 | 5.3 × 107 | 2.5 × 108 |
| 3—ProNaCC4 | 1.1 × 108 | 2.9 × 108 |
| 4—ProNaCC5 | 9.2 × 107 | 5.6 × 108 |
| 5—ProNaCC6 | 9.2 × 107 | 2.5 × 108 |
| 6—ProNaCC7 | 3.8 × 107 | 3.2 × 108 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macori, G.; Bellio, A.; Bianchi, D.M.; Chiesa, F.; Gallina, S.; Romano, A.; Zuccon, F.; Cabrera-Rubio, R.; Cauquil, A.; Merda, D.; et al. Genome-Wide Profiling of Enterotoxigenic Staphylococcus aureus Strains Used for the Production of Naturally Contaminated Cheeses. Genes 2020, 11, 33. https://doi.org/10.3390/genes11010033
Macori G, Bellio A, Bianchi DM, Chiesa F, Gallina S, Romano A, Zuccon F, Cabrera-Rubio R, Cauquil A, Merda D, et al. Genome-Wide Profiling of Enterotoxigenic Staphylococcus aureus Strains Used for the Production of Naturally Contaminated Cheeses. Genes. 2020; 11(1):33. https://doi.org/10.3390/genes11010033
Chicago/Turabian StyleMacori, Guerrino, Alberto Bellio, Daniela Manila Bianchi, Francesco Chiesa, Silvia Gallina, Angelo Romano, Fabio Zuccon, Raúl Cabrera-Rubio, Alexandra Cauquil, Déborah Merda, and et al. 2020. "Genome-Wide Profiling of Enterotoxigenic Staphylococcus aureus Strains Used for the Production of Naturally Contaminated Cheeses" Genes 11, no. 1: 33. https://doi.org/10.3390/genes11010033
APA StyleMacori, G., Bellio, A., Bianchi, D. M., Chiesa, F., Gallina, S., Romano, A., Zuccon, F., Cabrera-Rubio, R., Cauquil, A., Merda, D., Auvray, F., & Decastelli, L. (2020). Genome-Wide Profiling of Enterotoxigenic Staphylococcus aureus Strains Used for the Production of Naturally Contaminated Cheeses. Genes, 11(1), 33. https://doi.org/10.3390/genes11010033







