Inference of Breed Structure in Farm Animals: Empirical Comparison between SNP and Microsatellite Performance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. Microsatellites Datasets
2.1.2. SNPs Datasets
2.2. Data Analysis
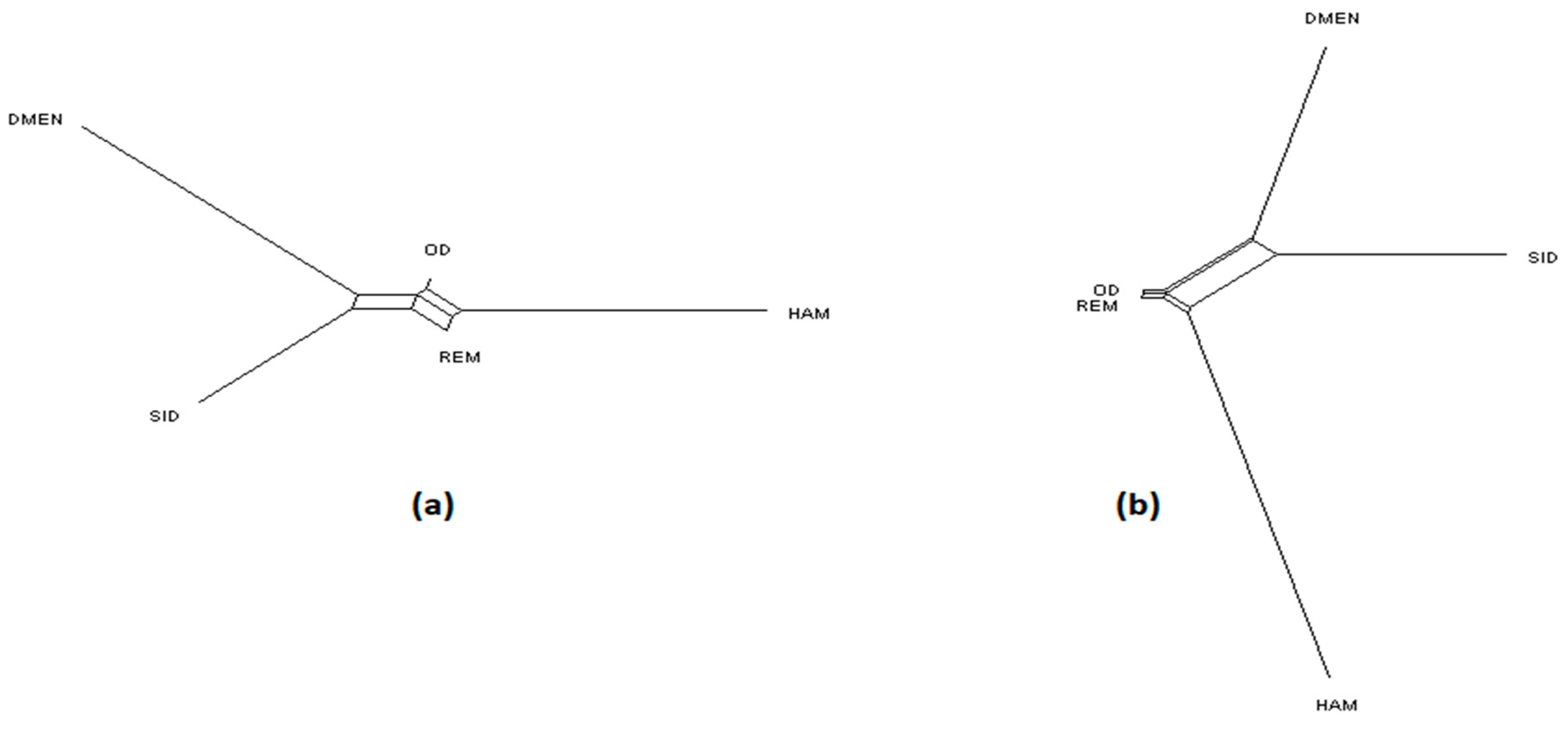
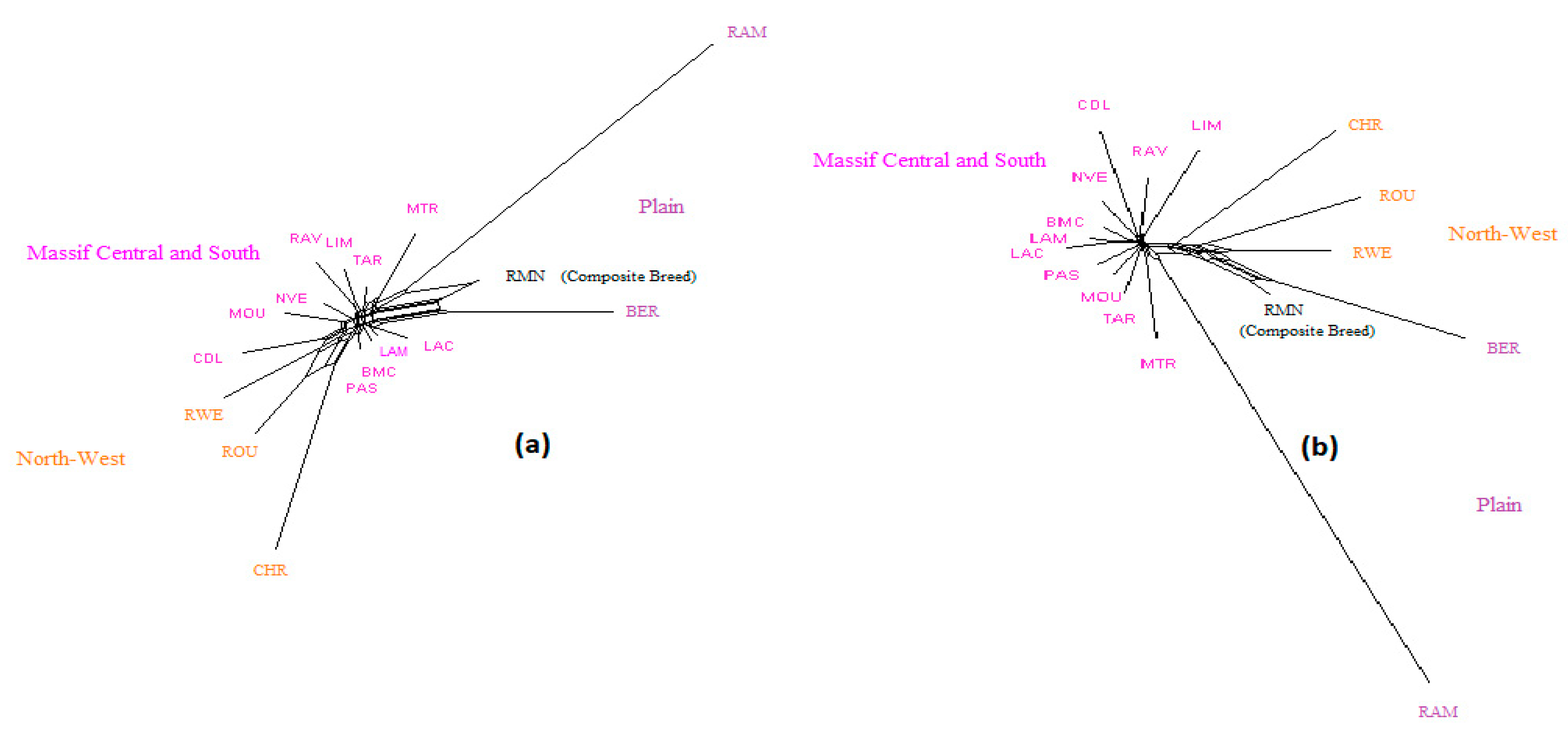
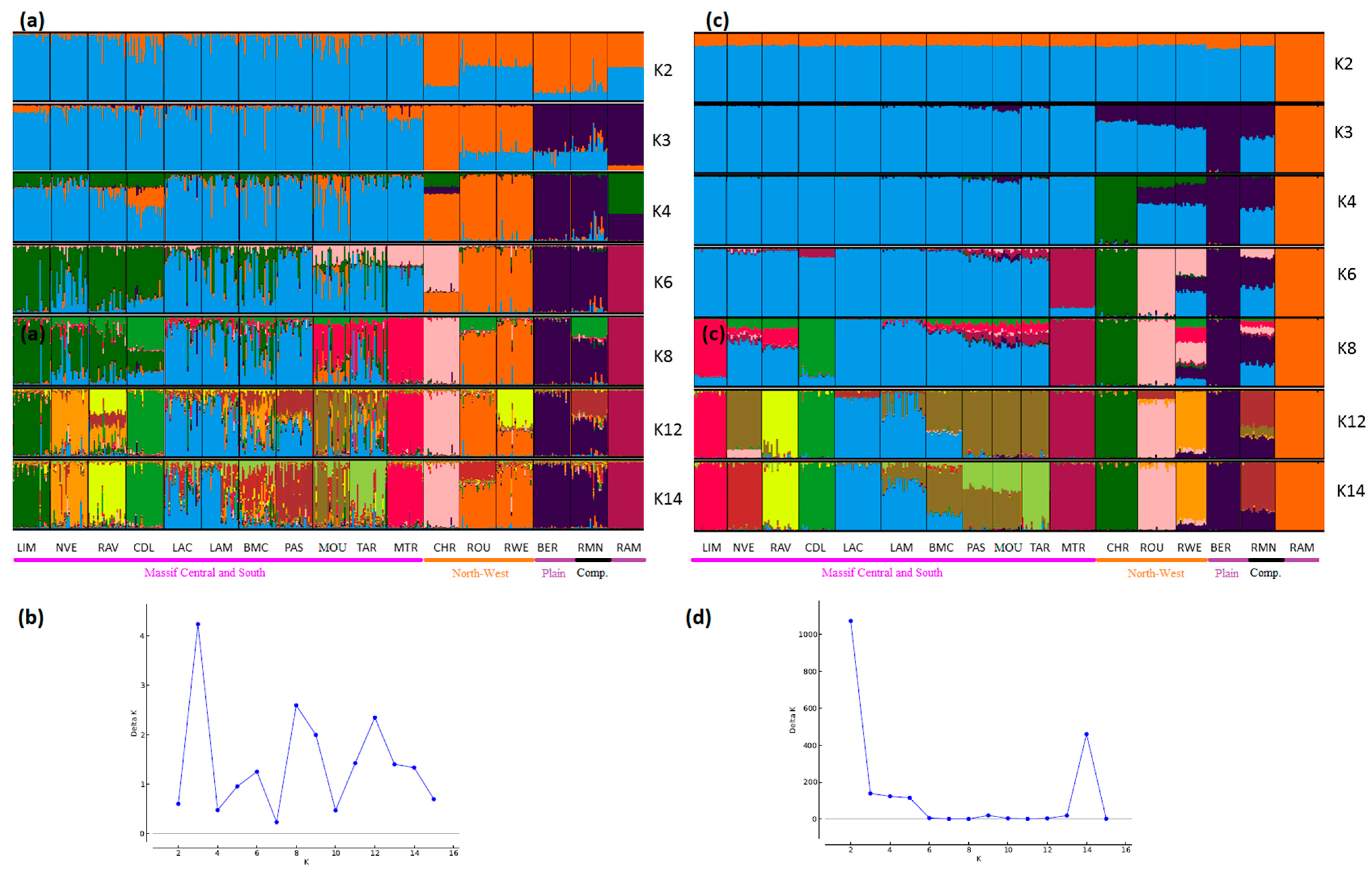
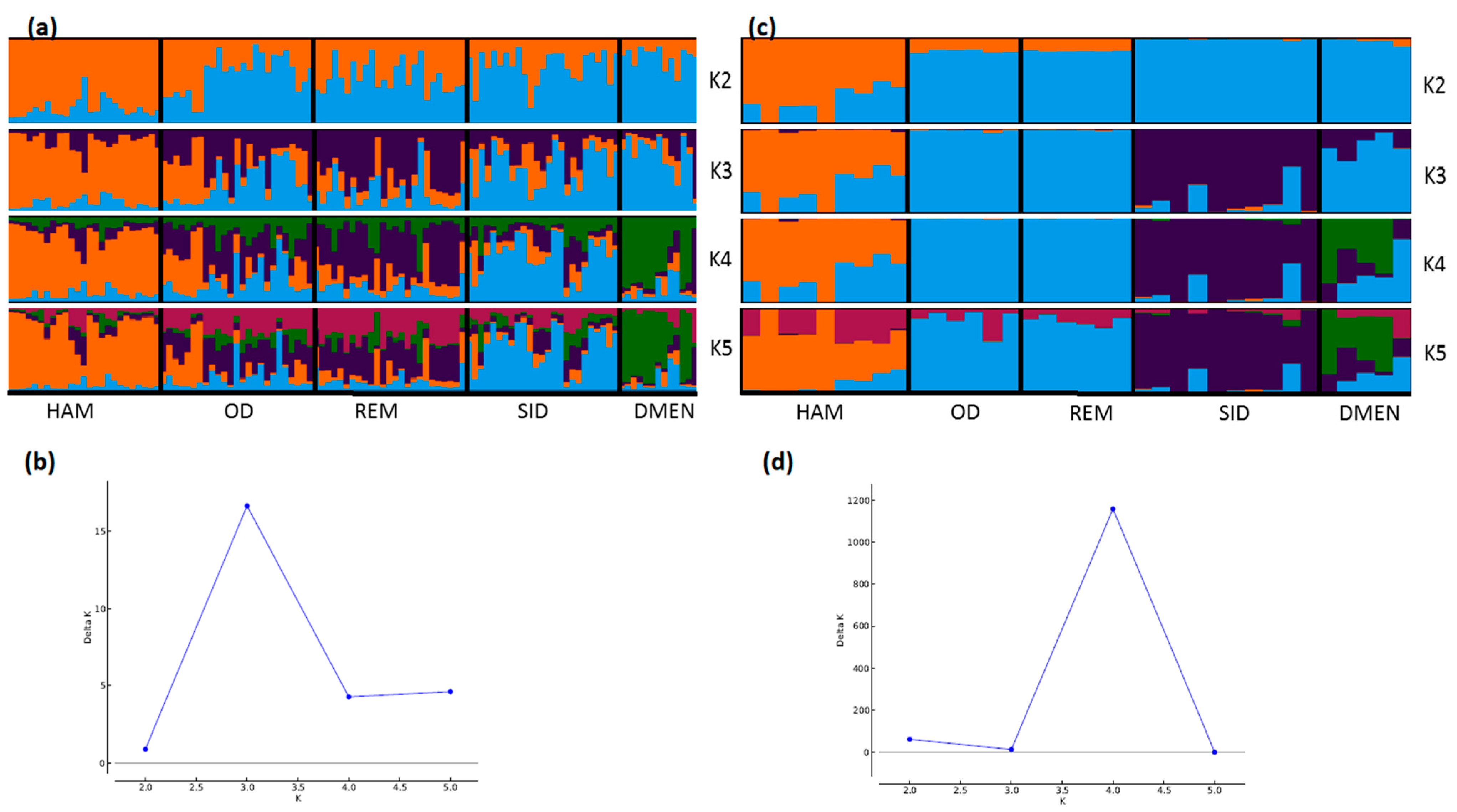
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Scherf, B.D., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2015; Available online: http://www.fao.org/3/a-i4787e/index.html (accessed on 23 September 2019).
- Sponenberg, D.P.; Beranger, J.; Martin, A. An Introduction to Heritage Breeds: Saving and Raising Rare-Breed Livestock and Poultry; Storey Publishing: North Adams, MA, USA, 2014. [Google Scholar]
- Bruford, M.W.; Ginja, C.; Hoffmann, I.; Joost, S.; Orozco-terWengel, P.; Alberto, F.J.; Amaral, A.J.; Barbato, M.; Biscarini, F.; Colli, L.; et al. Prospects and challenges for the conservation of farm animal genomic resources, 2015–2025. Front. Genet. 2015, 6, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magee, D.A.; Park, S.D.; Scraggs, E.; Murphy, A.M.; Doherty, M.L.; Kijas, J.W.; MacHugh, D.E. Technical note: High fidelity of whole-genome amplified sheep (Ovis aries) deoxyribonucleic acid using a high-density single nucleotide polymorphism array-based genotyping platform. J. Anim. Sci. 2010, 88, 3183–3186. [Google Scholar] [CrossRef] [Green Version]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.M.; Jamli, S.; et al. Design and Characterization of a 52K SNP Chip for Goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef] [PubMed]
- Groenen, M.A.; Megens, H.-J.; Zare, Y.; Warren, W.C.; Hillier, L.W.; Crooijmans, R.P.M.A.; Vereijken, A.; Okimoto, R.; Muir, W.M.; Cheng, H.H. The development and characterization of a 60K SNP chip for chicken. BMC Genom. 2011, 12, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, A.M.; Crooijmans, R.P.M.A.; Affara, N.A.; Amaral, A.J.; Archibald, A.L.; Beever, J.E.; Bendixen, C.; Churcher, C.; Clark, R.; Dehais, P.; et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE 2009, 4, e6524. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, S.A.; Taylor, S.A.; Chen, N.; Sillett, T.S.; Bondra, E.R.; Webster, M.S. A comparative assessment of SNP and microsatellite markers for assigning parentage in a socially monogamous bird. Mol. Ecol. Resour. 2017, 17, 183–193. [Google Scholar] [CrossRef]
- Trong, T.Q.; Bers, v.N.; Crooijmans, R.P.M.A.; Dibbits, B.W.; Komen, J. A comparison of microsatellites and SNPs in parental assignment in the GIFT strain of Nile tilapia (Oreochromis niloticus): The power of exclusion. Aquaculture 2013, 388–391, 14–23. [Google Scholar] [CrossRef]
- Weinman, L.R.; Solomon, J.W.; Rubenstein, D.R. A comparison of single nucleotide polymorphism and microsatellite markers for analysis of parentage and kinship in a cooperatively breeding bird. Mol. Ecol. Resour. 2015, 15, 502–511. [Google Scholar] [CrossRef]
- Fernández, M.E.; Goszczynski, D.E.; Lirón, J.P.; Villegas-Castagnasso, E.E.; Carino, M.H.; Ripoli, M.V.; Rogberg-Muoz, A.; Posik, D.M.; Peral-Garca, P.; Giovambattista, G. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd. Genet. Mol. Biol. 2013, 36, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.C.; Tang, Q.Z.; Long, K.R.; Che, T.D.; Li, M.Z.; Shuai, S.R. Effectiveness of microsatellite and single nucleotide polymorphism markers for parentage analysis in European domestic pigs. Genet. Mol. Res. 2015, 14, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Cortés, O.; Eusebi, P.; Dunner, S.; Sevane, N.; Cañón, J. Comparison of diversity parameters from SNP, microsatellites and pedigree records in the Lidia cattle breed. Livest. Sci. 2019, 219, 80–85. [Google Scholar] [CrossRef]
- Ljungqvist, M.; Akesson, M.; Hansson, B. Do microsatellites reflect genome-wide genetic diversity in natural populations? A comment on Väli et al. (2008). Mol. Ecol. 2010, 19, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.A.; Archer, F.I.; Pease, V.L.; Hancock-Hanser, B.L.; Robertson, K.M.; Huebinger, R.M.; Martien, K.K.; Bickham, J.W.; George, J.C.; Postma, L.D.; et al. Empirical comparison of single nucleotide polymorphisms and microsatellites for population and demographic analyses of bowhead whales. Endanger. Spec. Res. 2012, 19, 129–147. [Google Scholar] [CrossRef]
- Glover, K.A.; Hansen, M.M.; Lien, S.; Als, T.D.; Høyheim, B.; Skaala, Ø. A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genet. 2010, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Gärke, C.; Ytournel, F.; Bed’hom, B.; Gut, I.; Lathrop, M.; Weigend, S.; Simianer, H. Comparison of SNPs and microsatellites for assessing the genetic structure of chicken populations. Anim. Genet. 2012, 43, 419–428. [Google Scholar] [CrossRef]
- Saint-Pé, K.; Leitwein, M.; Tissot, L.; Poulet, N.; Guinand, B.; Berrebi, P.; Marselli, G.; Lascaux, J.-M.; Gagnaire, P.-A.; Blanchet, S. Development of a large SNPs resource and a low-density SNP array for brown trout (Salmo trutta) population genetics. BMC Genom. 2019, 20, 582. [Google Scholar] [CrossRef] [Green Version]
- Ciani, E.; Cecchi, F.; Castellana, E.; D’Andrea, M.; Incoronato, C.; D’Angelo, F.; Albenzio, M.; Pilla, F.; Matassino, D.; Cianci, D.; et al. Poorer resolution of low-density SNP vs. STR markers in reconstructing genetic relationships among seven Italian sheep breeds. Large Anim. Rev. 2013, 19, 236–241. [Google Scholar]
- Leroy, G.; Danchin-Burge, C.; Palhière, I.; SanCristobal, M.; Nédélec, Y.; Verrier, E.; Rognon, X. How do introgression events shape the partitioning of diversity among breeds: A case study in sheep. Genet. Sel. Evol. 2015, 47, 48. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Darly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- FAO, Food and Agricultural Organization of the United Nations. Molecular Genetic Characterization of Animal Genetic Resources; FAO Animal Production and Health Guidelines: Rome, Italy, 2011; pp. 65–84. [Google Scholar]
- FAO, Food and Agriculture Organization of the United Nations. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans. In Measurement of Domestic Animal Diversity (MoDAD): Recommended Microsatellite Markers; Available online: http://dad.fao.org/en/refer/library/guidelin/marker.pdf (accessed on 23 September 2019).
- Gaouar, S.B.S.; Da Silva, A.; Ciani, E.; Kdidi, S.; Aouissat, M.; Dhimi, L.; Lafri, M.; Maftah, A.; Mehtar, N. Admixture and Local Breed Marginalization Threaten Algerian Sheep Diversity. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, J.P.; Royo, L.J.; Álvarez, I.; Goyache, F. MolKin v2.0: A computer program for genetic analysis of populations using molecular coancestry information. J. Hered. 2005, 96, 718–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sempéré, G.; Moazami-Goudarzi, K.; Eggen, A.; Laloë, D.; Gautier, M.; Flori, L. WIDDE: A Web-Interfaced next generation database for genetic diversity exploration, with a first application in cattle. BMC Genom. 2015, 16, 940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaouar, S.B.S.; Lafri, M.; Djaout, A.; El-Bouyahiaoui, R.; Bouri, A.; Bouchatal, A.; Maftah, A.; Ciani, E.; Da Silva, A.B. Genome-wide analysis highlights genetic dilution in Algerian sheep. Heredity 2017, 118, 293–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belabdi, I.; Ouhrouch, A.; Lafri, M.; Gaouar, S.B.S.; Ciani, E.; Benali, A.R.; Ouelhadj, H.O.; Haddioui, A.; Pompanon, F.; Blanquet, V.; et al. Genetic homogenization of indigenous sheep breeds in Northwest Africa. Sci. Rep. 2019, 9, 7920. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Romieux, C.; Tortereau, F.; Raoul, J.; Servin, B. High density genotypes of French Sheep populations [Internet]. Zenodo 2017. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2006. [Google Scholar]
- Malomane, D.K.; Reimer, C.; Weigend, S.; Weigend, A.; Sharifi, A.R.; Simianer, H. Efficiency of different strategies to mitigate ascertainment bias when using SNP panels in diversity studies. BMC Genom. 2018, 19, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, J.; Weir, B.S.; Cockerham, C. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics 1983, 105, 767–779. [Google Scholar] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–627. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Ducrocq, V.; Laloe, D.; Sawaminathan, M.; Rognon, X.; Tixier-Boichard, M.; Zerjal, T. Genomics for Ruminants in Developing Countries: From Principles to Practice. Front. Genet. 2018, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Leroy, G.; Kayang, B.B.; Youssao, I.A.K.; Yapi-Gnaore, C.V.; Osei-Amponsah, R.; Loukou, N.E.; Fotsa, J.C.; Benabdeljelil, K.; Bed’hom, B.; Tixier-Boichard, M.; et al. 2012 Gene diversity, agroecological structure and introgression patterns among village chicken populations across north, west and central Africa. BMC Genet. 2012, 13, 34. [Google Scholar] [CrossRef] [Green Version]
- Harkat, S.; Laoun, A.; Belabdi, I.; Benali, R.; Outayeb, D.; Payet-Duprat, N.; Blanquet, V.; Lafri, M.; Da Silva, A. Assessing patterns of genetic admixture between sheep breeds: Case study in Algeria. Ecol Evol. 2017, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selepe, M.M.; Ceccobelli, S.; Lasagna, E.; Kunene, N.W. Genetic structure of South African Nguni (Zulu) sheep populations reveals admixture with exotic breeds. PLoS ONE 2018, 13, e0196276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taberlet, P.; Valentini, A.; Rezaei, H.R.; Naderi, S.; Pompanon, F.; Negrini, R.; Ajmone-Marsan, P. Are cattle, sheep, and goats endangered species? Mol. Ecol. 2008, 17, 275284. [Google Scholar] [CrossRef] [PubMed]


| Algerian Sheep Datasets | French Sheep Datasets | French Cattle Datasets | |
|---|---|---|---|
| no. of breeds | 5 | 17 | 7 |
| Microsatellite datasets: | |||
| nb. of individuals | 113 | 425 | 175 |
| nb. of microsatellites | 29 | 21 | 30 |
| nb. of alleles | 343 | 292 | 283 |
| mean A (s.d.) | 11.83 (13.29) | 13.90 (21.59) | 9.43 (11.97) |
| mean HO (s.d.) | 0.77 (0.008) | 0.73 (0.018) | 0.71 (0.015) |
| mean PIC (s.d.) | 0.74 (0.010) | 0.70 (0.018) | 0.67 (0.018) |
| mean Ae (s.d.) | 5.06 (4.67) | 4.46 (3.89) | 3.98 (3.02) |
| SNP datasets: | |||
| nb. of individuals * | 36 | 346 | 152 |
| nb. of SNP | 52,412 | 40,454 | 52,324 |
| nb. of SNP after filtration | 36,493 | 39,800 | 47,286 |
| nb. of SNP after Pruning ** | 15,560 | 31,184 | 24,841 |
| Mean FST from microsatellites datasets (s.d.) | 0.048 (<0.001) | 0.104 (0.004) | 0.076 (<0.001) |
| Mean FST from SNP datasets (s.d.) | 0.048 (<0.001) | 0.105 (0.004) | 0.078 (<0.001) |
| r Pearson *** (p-value) | 0.87 (0.001) | 0.95 (<0.001) | 0.77 (<0.001) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laoun, A.; Harkat, S.; Lafri, M.; Gaouar, S.B.S.; Belabdi, I.; Ciani, E.; De Groot, M.; Blanquet, V.; Leroy, G.; Rognon, X.; et al. Inference of Breed Structure in Farm Animals: Empirical Comparison between SNP and Microsatellite Performance. Genes 2020, 11, 57. https://doi.org/10.3390/genes11010057
Laoun A, Harkat S, Lafri M, Gaouar SBS, Belabdi I, Ciani E, De Groot M, Blanquet V, Leroy G, Rognon X, et al. Inference of Breed Structure in Farm Animals: Empirical Comparison between SNP and Microsatellite Performance. Genes. 2020; 11(1):57. https://doi.org/10.3390/genes11010057
Chicago/Turabian StyleLaoun, Abbas, Sahraoui Harkat, Mohamed Lafri, Semir Bechir Suheil Gaouar, Ibrahim Belabdi, Elena Ciani, Maarten De Groot, Véronique Blanquet, Gregoire Leroy, Xavier Rognon, and et al. 2020. "Inference of Breed Structure in Farm Animals: Empirical Comparison between SNP and Microsatellite Performance" Genes 11, no. 1: 57. https://doi.org/10.3390/genes11010057
APA StyleLaoun, A., Harkat, S., Lafri, M., Gaouar, S. B. S., Belabdi, I., Ciani, E., De Groot, M., Blanquet, V., Leroy, G., Rognon, X., & Da Silva, A. (2020). Inference of Breed Structure in Farm Animals: Empirical Comparison between SNP and Microsatellite Performance. Genes, 11(1), 57. https://doi.org/10.3390/genes11010057





