Glabralysins, Potential New β-Pore-Forming Toxin Family Members from the Schistosomiasis Vector Snail Biomphalaria glabrata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethic Statement
2.2. Biological Material
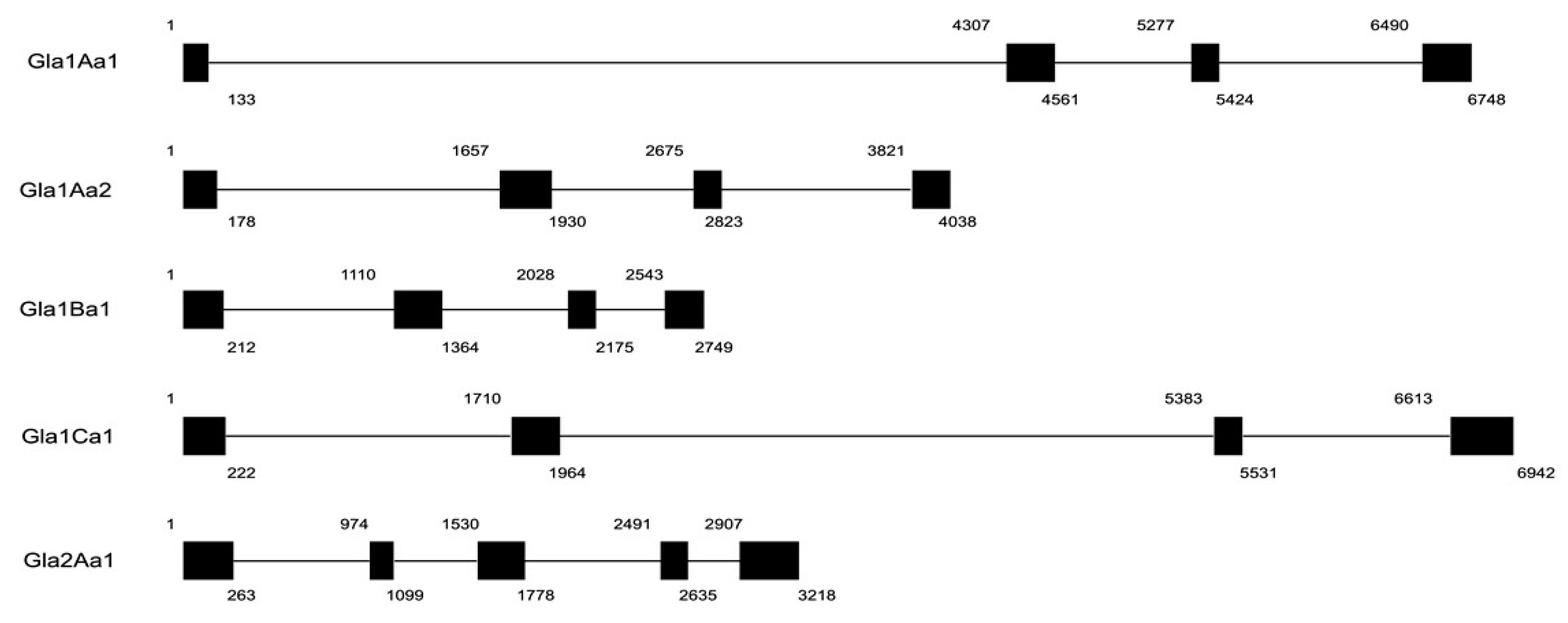
2.3. Characterization of Glabralysin Gene Organization
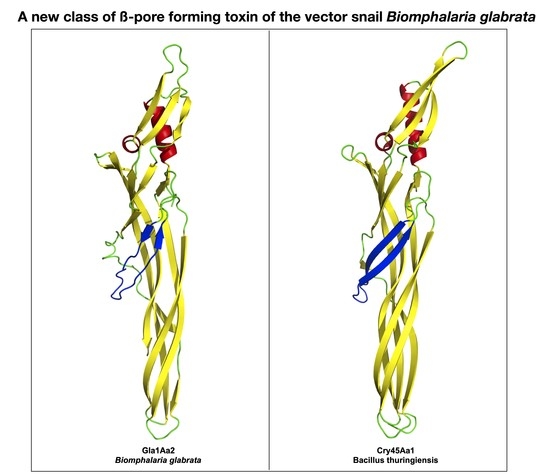
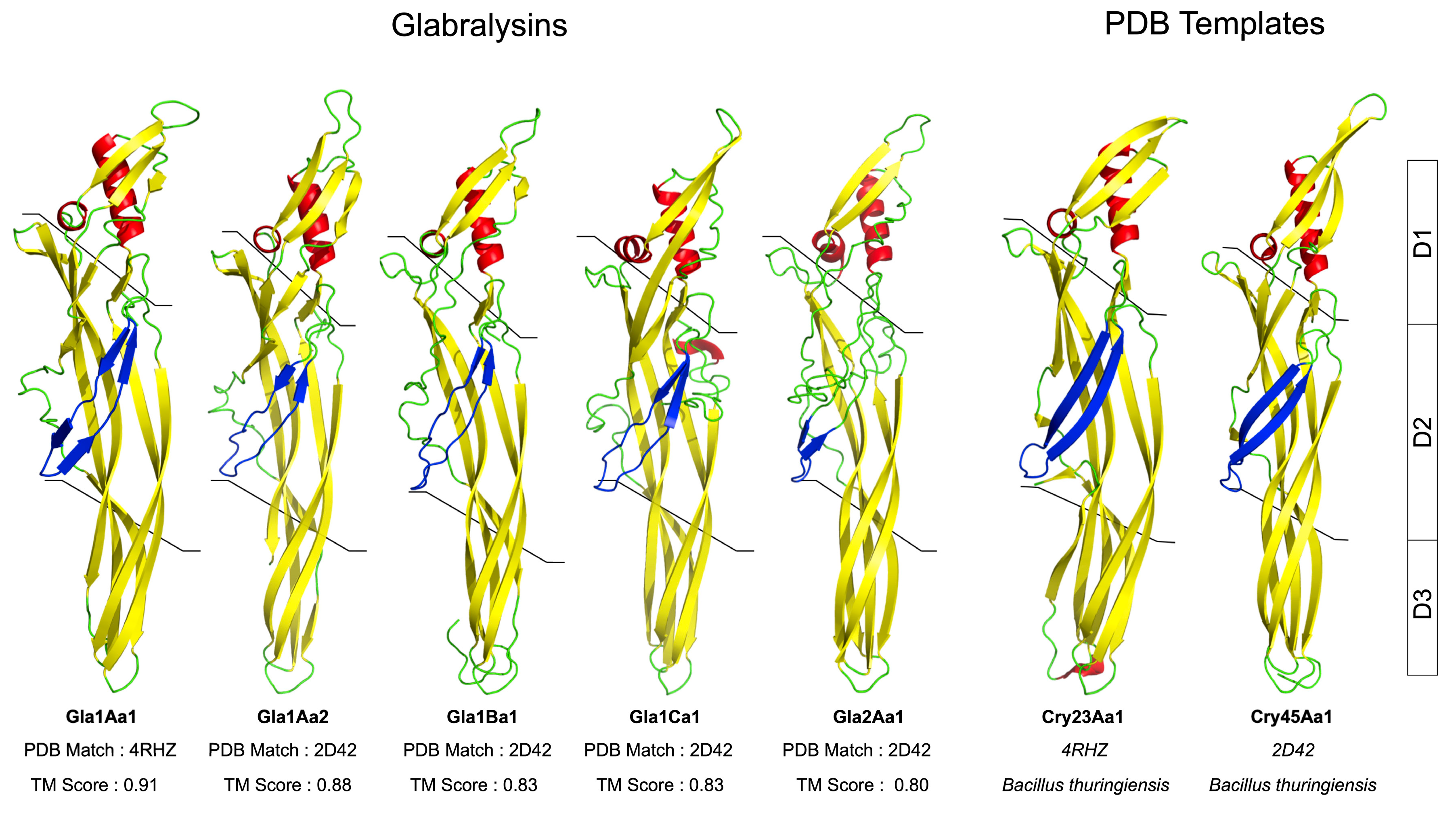
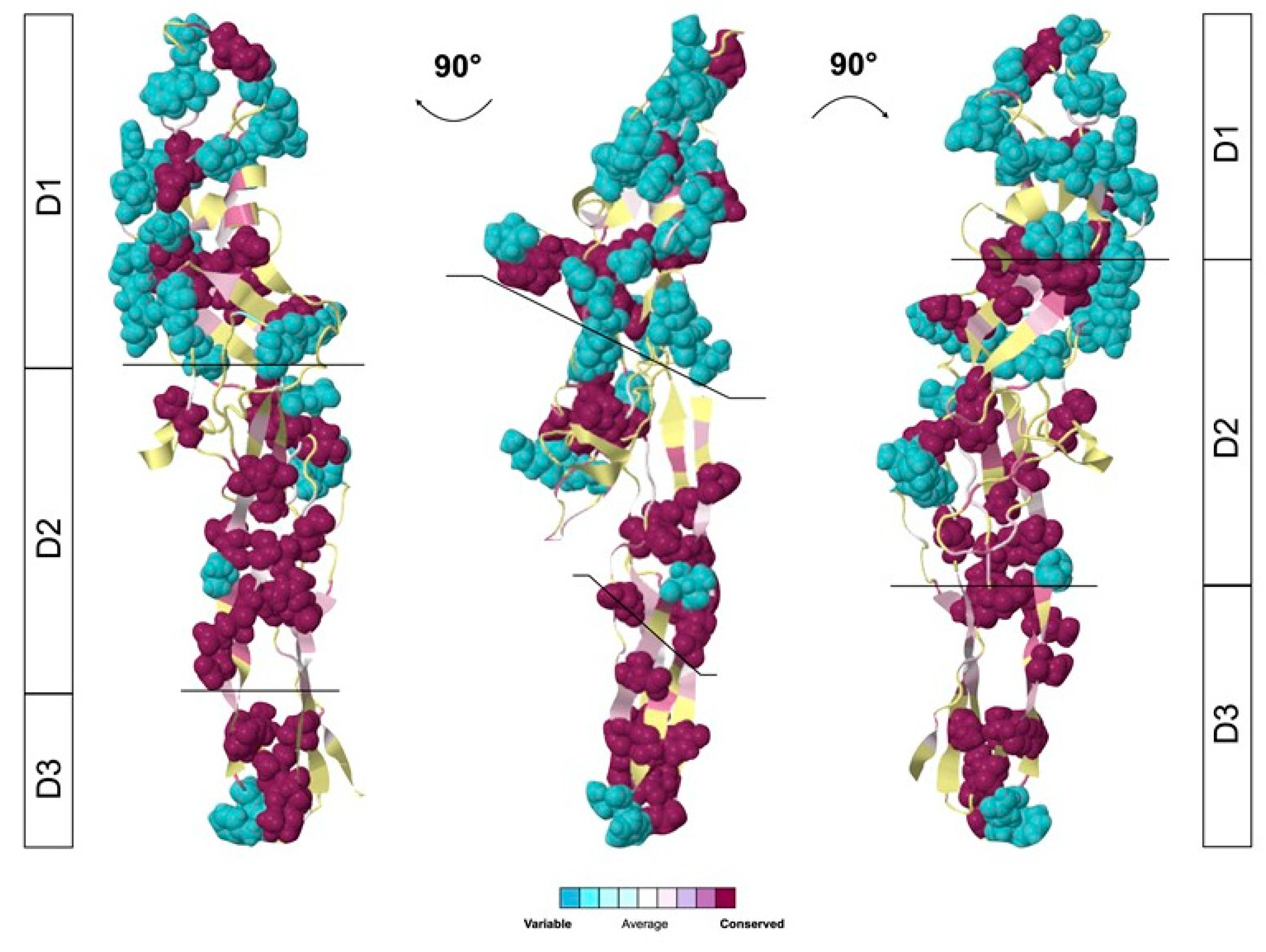
2.4. Glabralysin 3D Structure Prediction and Analysis
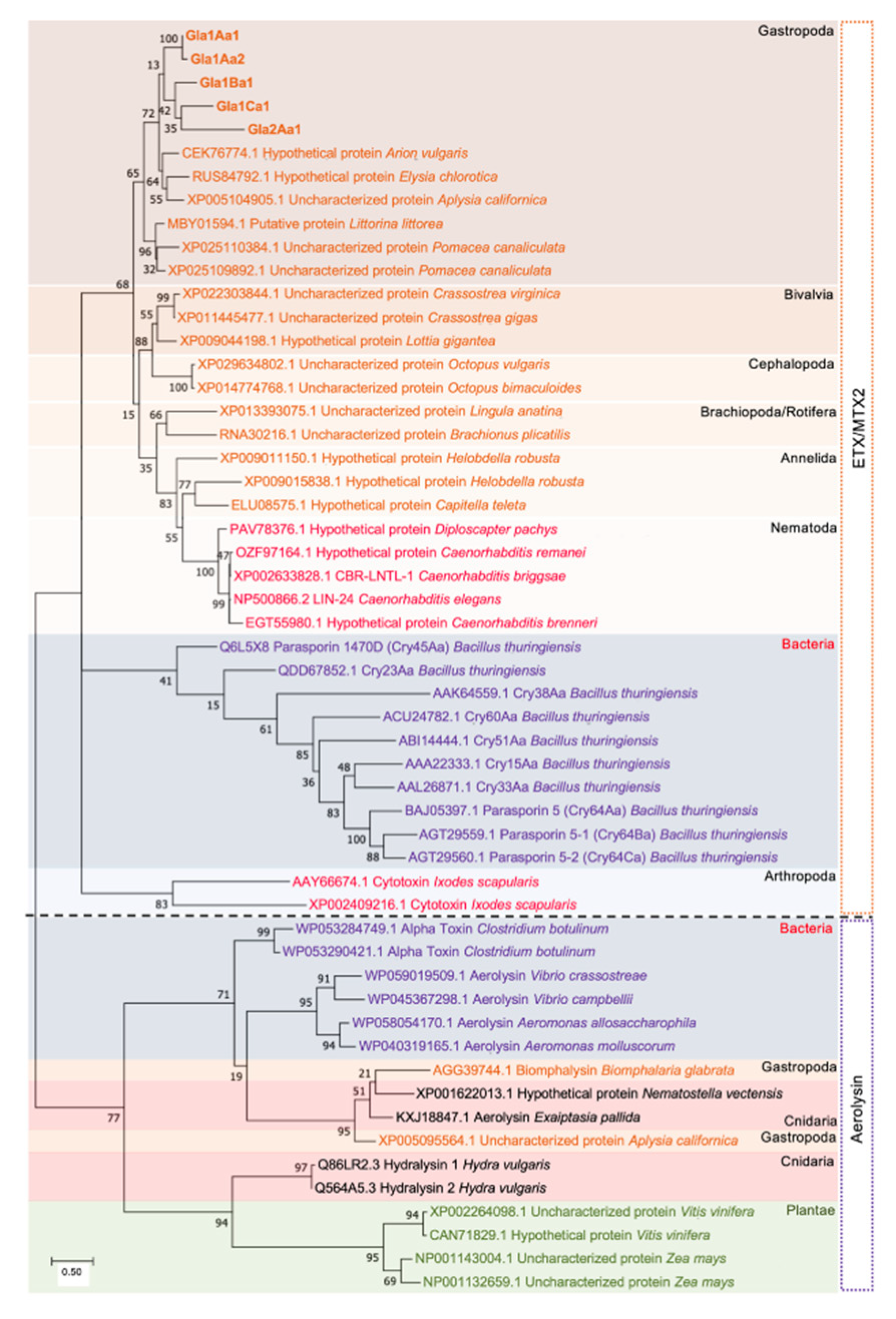
2.5. Phylogenetic Tree Analysis
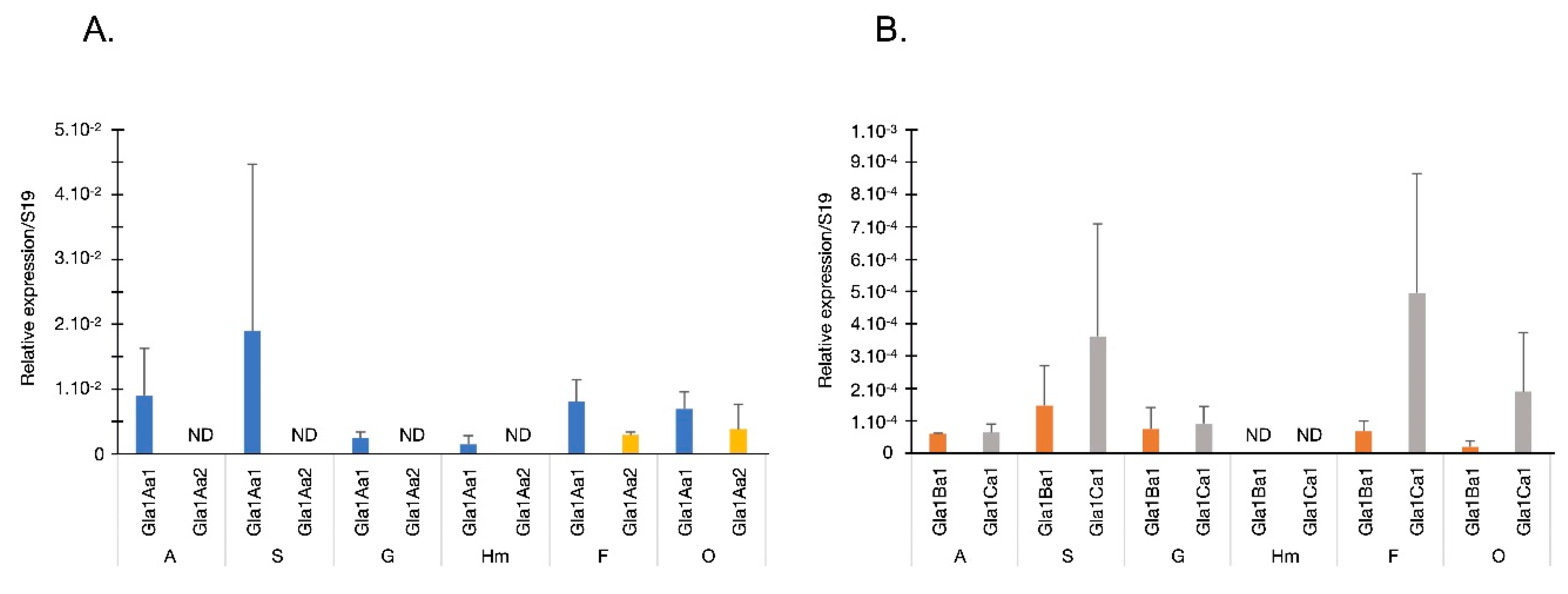
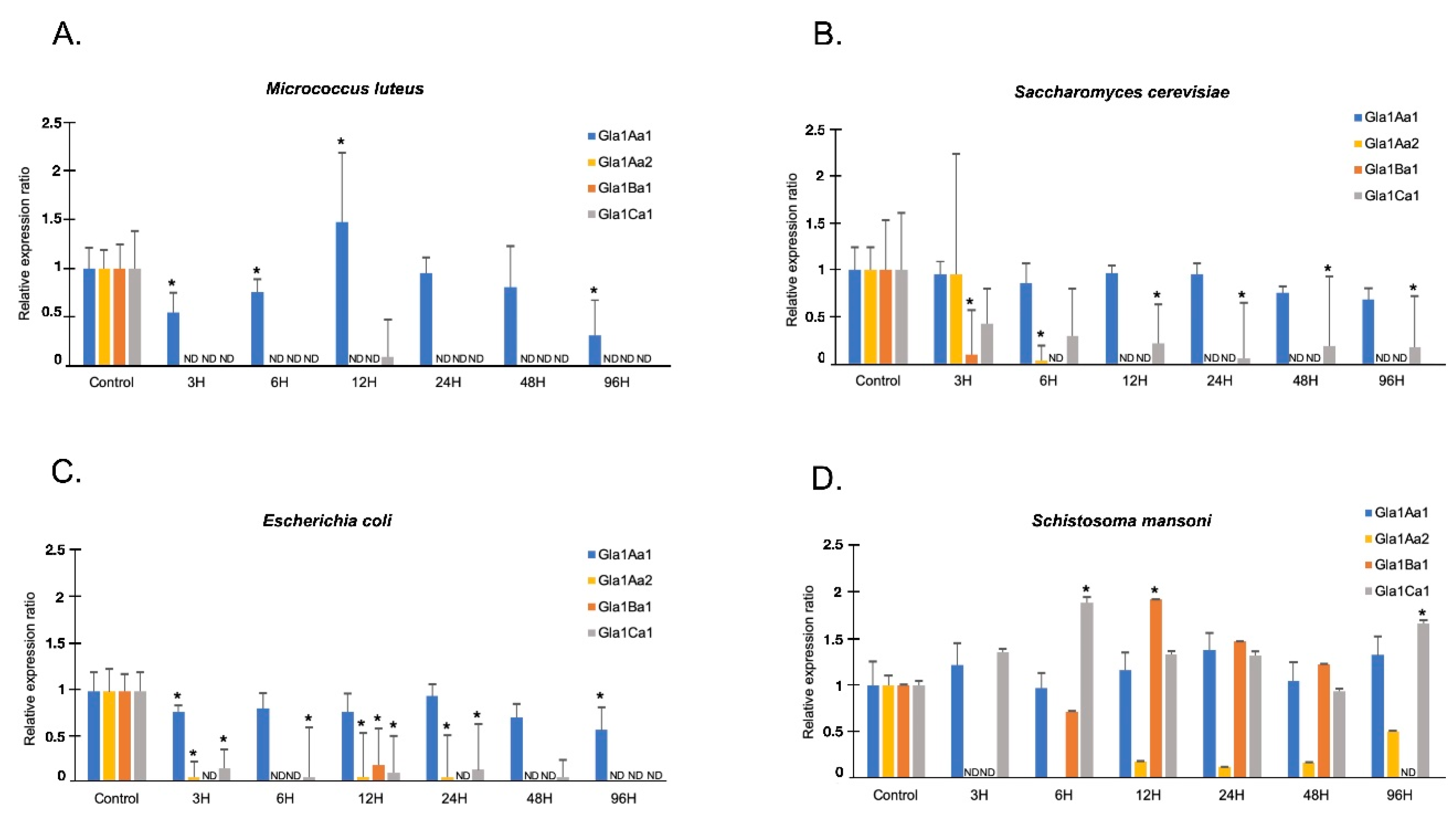
2.6. Quantitative Real Time PCR Assay for Glabralysins Expression Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Combes, C. Selective pressure in host-parasite systems. J. Soc. Biol. 2000, 194, 19–23. [Google Scholar] [CrossRef]
- Horrocks, N.P.C.; Matson, K.D.; Tieleman, B.I. Pathogen pressure puts immune defense into perspective. Integr. Comp. Biol. 2011, 51, 563–576. [Google Scholar] [CrossRef]
- Janeway, C.A.; Bottomly, K. Signals and signs for lymphocyte responses. Cell 1994, 76, 275–285. [Google Scholar] [CrossRef]
- Coustau, C.; Gourbal, B.; Duval, D.; Yoshino, T.P.; Adema, C.M.; Mitta, G. Advances in gastropod immunity from the study of the interaction between the snail Biomphalaria glabrata and its parasites: A review of research progress over the last decade. Fish Shellfish Immunol. 2015, 46, 5–16. [Google Scholar]
- Wang, J.; Song, X.; Wang, M. Peptidoglycan recognition proteins in hematophagous arthropods. Dev. Comp. Immunol. 2018, 83, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S. Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 2017, 38, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Cot, M.; Puzo, G.; Gilleron, M.; Nigou, J. Bacterial cell wall macroamphiphiles: Pathogen-/microbe-associated molecular patterns detected by mammalian innate immune system. Biochimie 2013, 95, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Galinier, R.; Tetreau, G.; Portet, A.; Pinaud, S.; Duval, D.; Gourbal, B. First characterization of viruses from freshwater snails of the genus Biomphalaria, the intermediate host of the parasite Schistosoma mansoni. Acta Trop. 2017, 167, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Duval, D.; Galinier, R.; Mouahid, G.; Toulza, E.; Allienne, J.F.; Portela, J.; Calvayrac, C.; Rognon, A.; Arancibia, N.; Mitta, G.; et al. A novel bacterial pathogen of Biomphalaria glabrata: a potential weapon for schistosomiasis control? PLoS Negl. Trop. Dis. 2015, 9, e0003489. [Google Scholar]
- Hertel, L.A.; Barbosa, C.S.; Santos, R.A.A.L.; Loker, E.S. Molecular identification of symbiont from the pulmonate snail Biomphalaria glabrata in Brazil. J. Parasitol. 2004, 90, 759–763. [Google Scholar] [CrossRef]
- Baron, O.L.; van West, P.; Industri, B.; Ponchet, M.; Dubreuil, G.; Gourbal, B.; Reichhart, J.-M.; Coustau, C. Parental transfer of the antimicrobial protein LBP/BPI protects Biomphalaria glabrata eggs against oomycete infections. PLoS Pathog. 2013, 9, e1003792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousif, F.; Lämmler, G. The mode of infection with and the distribution of Angiostrongylus cantonensis larvae in the experimental intermediate hoist Biomphalaria glabrata. Z. Parasitenkd. 1977, 53, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.T. Experimental second intermediate hosts of Echinostoma malayanum Leiper, 1911. J. Parasitol. 1973, 59, 746–747. [Google Scholar] [CrossRef] [PubMed]
- Goodall, C.P.; Bender, R.C.; Brooks, J.K.; Bayne, C.J. Biomphalaria glabrata cytosolic copper/zinc superoxide dismutase (SOD1) gene: association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni. Mol. Biochem. Parasitol. 2006, 147, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Bonner, K.M.; Bayne, C.J.; Larson, M.K.; Blouin, M.S. Effects of Cu/Zn superoxide dismutase (sod1) genotype and genetic background on growth, reproduction and defense in Biomphalaria glabrata. PLoS Negl. Trop. Dis. 2012, 6, e1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tennessen, J.A.; Théron, A.; Marine, M.; Yeh, J.-Y.; Rognon, A.; Blouin, M.S. Hyperdiverse gene cluster in snail host conveys resistance to human schistosome parasites. PLoS Genet. 2015, 11, e1005067. [Google Scholar] [CrossRef] [Green Version]
- Tennessen, J.A.; Bonner, K.M.; Bollmann, S.R.; Johnstun, J.A.; Yeh, J.-Y.; Marine, M.; Tavalire, H.F.; Bayne, C.J.; Blouin, M.S. Genome-Wide Scan and Test of Candidate Genes in the Snail Biomphalaria glabrata Reveal New Locus Influencing Resistance to Schistosoma mansoni. PLoS Negl. Trop. Dis. 2015, 9, e0004077. [Google Scholar] [CrossRef]
- Lockyer, A.E.; Noble, L.R.; Rollinson, D.; Jones, C.S. Schistosoma mansoni: resistant specific infection-induced gene expression in Biomphalaria glabrata identified by fluorescent-based differential display. Exp. Parasitol. 2004, 107, 97–104. [Google Scholar] [CrossRef]
- Lockyer, A.E.; Spinks, J.N.; Walker, A.J.; Kane, R.A.; Noble, L.R.; Rollinson, D.; Dias-Neto, E.; Jones, C.S. Biomphalaria glabrata transcriptome: Identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES). Dev. Comp. Immunol. 2007, 31, 763–782. [Google Scholar] [CrossRef] [Green Version]
- Galinier, R.; Roger, E.; Moné, Y.; Duval, D.; Portet, A.; Pinaud, S.; Chaparro, C.; Grunau, C.; Genthon, C.; Dubois, E.; et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017, 11, e0005398. [Google Scholar] [CrossRef]
- Bayne, C.J.; Hahn, U.K.; Bender, R.C. Mechanisms of molluscan host resistance and of parasite strategies for survival. Parasitology 2001, 123, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.; Ittiprasert, W.; Raghavan, N.; Miller, A.; Knight, M. Differences in cysteine protease activity in Schistosoma mansoni-resistant and -susceptible Biomphalaria glabrata and characterization of the hepatopancreas cathepsin B Full-length cDNA. J. Parasitol. 2008, 94, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Roger, E.; Grunau, C.; Pierce, R.J.; Hirai, H.; Gourbal, B.; Galinier, R.; Emans, R.; Cesari, I.M.; Cosseau, C.; Mitta, G. Controlled Chaos of Polymorphic Mucins in a Metazoan Parasite (Schistosoma mansoni) Interacting with Its Invertebrate Host (Biomphalaria glabrata). PLoS Negl. Trop. Dis. 2008, 2, e330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moné, Y.; Gourbal, B.; Duval, D.; Du Pasquier, L.; Kieffer-Jaquinod, S.; Mitta, G. A Large Repertoire of Parasite Epitopes Matched by a Large Repertoire of Host Immune Receptors in an Invertebrate Host/Parasite Model. PLoS Negl. Trop. Dis. 2010, 4, e813. [Google Scholar] [CrossRef] [Green Version]
- Galinier, R.; Portela, J.; Moné, Y.; Allienne, J.F.; Henri, H.; Delbecq, S.; Mitta, G.; Gourbal, B.; Duval, D. Biomphalysin, a New β Pore-forming Toxin Involved in Biomphalaria glabrata Immune Defense against Schistosoma mansoni. PLoS Pathog. 2013, 9, e1003216. [Google Scholar] [CrossRef]
- Pila, E.A.; Gordy, M.A.; Phillips, V.K.; Kabore, A.L.; Rudko, S.P.; Hanington, P.C. Endogenous growth factor stimulation of hemocyte proliferation induces resistance to Schistosoma mansoni challenge in the snail host. Proc. Natl. Acad. Sci. USA 2016, 113, 5305–5310. [Google Scholar] [CrossRef] [Green Version]
- Tetreau, G.; Pinaud, S.; Portet, A.; Galinier, R.; Gourbal, B.; Duval, D. Specific Pathogen Recognition by Multiple Innate Immune Sensors in an Invertebrate. Front. Immunol. 2017, 8, 1249. [Google Scholar] [CrossRef]
- Ittiprasert, W.; Nene, R.; Miller, A.; Raghavan, N.; Lewis, F.; Hodgson, J.; Knight, M. Schistosoma mansoni infection of juvenile Biomphalaria glabrata induces a differential stress response between resistant and susceptible snails. Exp. Parasitol. 2009, 123, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Ittiprasert, W.; Knight, M. Reversing the Resistance Phenotype of the Biomphalaria glabrata Snail Host Schistosoma mansoni Infection by Temperature Modulation. PLoS Pathog. 2012, 8, e1002677. [Google Scholar] [CrossRef]
- Lepesant, J.M.J.; Cosseau, C.; Boissier, J.; Freitag, M.; Portela, J.; Climent, D.; Perrin, C.; Zerlotini, A.; Grunau, C. Chromatin structural changes around satellite repeats on the female sex chromosome in Schistosoma mansoni and their possible role in sex chromosome emergence. Genome Biol. 2012, 13, R14. [Google Scholar] [CrossRef]
- Bridger, J.M.; Arican-Gotkas, H.D.; Foster, H.A.; Godwin, L.S.; Harvey, A.; Kill, I.R.; Knight, M.; Mehta, I.S.; Ahmed, M.H. The non-random repositioning of whole chromosomes and individual gene loci in interphase nuclei and its relevance in disease, infection, aging, and cancer. Adv. Exp. Med. Biol. 2014, 773, 263–279. [Google Scholar] [PubMed]
- Knight, M.; Elhelu, O.; Smith, M.; Haugen, B.; Miller, A.; Raghavan, N.; Wellman, C.; Cousin, C.; Dixon, F.; Mann, V.; et al. Susceptibility of Snails to Infection with Schistosomes is influenced by Temperature and Expression of Heat Shock Proteins. Epidemiology (Sunnyvale) 2015, 5, 189. [Google Scholar] [PubMed] [Green Version]
- Pila, E.A.; Tarrabain, M.; Kabore, A.L.; Hanington, P.C. A Novel Toll-Like Receptor (TLR) Influences Compatibility between the Gastropod Biomphalaria glabrata, and the Digenean Trematode Schistosoma mansoni. PLoS Pathog. 2016, 12, e1005513. [Google Scholar] [CrossRef] [Green Version]
- Humphries, J.E.; Deneckere, L.E. Characterization of a Toll-like receptor (TLR) signaling pathway in Biomphalaria glabrata and its potential regulation by NF-kappaB. Dev. Comp. Immunol. 2018, 86, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Dheilly, N.M.; Duval, D.; Mouahid, G.; Emans, R.; Allienne, J.-F.; Galinier, R.; Genthon, C.; Dubois, E.; Du Pasquier, L.; Adema, C.M.; et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev. Comp. Immunol. 2015, 48, 234–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.-J.; Dinguirard, N.; Sabat, G.; Lui, H.; Gonzalez, L.; Gehring, M.; Bickham-Wright, U.; Yoshino, T.P. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PLoS Pathog. 2017, 13, e1006081. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-M.; Loker, E.S. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Fibrinogen-related proteins. Dev. Comp. Immunol. 2003, 27, 175–187. [Google Scholar] [CrossRef]
- Ittiprasert, W.; Miller, A.; Myers, J.; Nene, V.; El-Sayed, N.M.; Knight, M. Identification of immediate response genes dominantly expressed in juvenile resistant and susceptible Biomphalaria glabrata snails upon exposure to Schistosoma mansoni. Mol. Biochem. Parasitol. 2010, 169, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Lockyer, A.E.; Emery, A.M.; Kane, R.A.; Walker, A.J.; Mayer, C.D.; Mitta, G.; Coustau, C.; Adema, C.M.; Hanelt, B.; Rollinson, D.; et al. Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni. PLoS ONE 2012, 7, e51102. [Google Scholar] [CrossRef]
- Mitta, G.; Galinier, R.; Tisseyre, P.; Allienne, J.-F.; Girerd-Chambaz, Y.; Guillou, F.; Bouchut, A.; Coustau, C. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev. Comp. Immunol. 2005, 29, 393–407. [Google Scholar] [CrossRef]
- Hahn, U.K.; Bender, R.C.; Bayne, C.J. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: role of reactive oxygen species. J. Parasitol. 2001, 87, 292–299. [Google Scholar] [CrossRef]
- de Moraes Mourão, M.; Dinguirard, N.; Franco, G.R.; Yoshino, T.P. Phenotypic Screen of Early-Developing Larvae of the Blood Fluke, Schistosoma mansoni, using RNA Interference. PLoS Negl. Trop. Dis. 2009, 3, e502. [Google Scholar] [CrossRef]
- Baron, O.L.; Deleury, E.; Reichhart, J.-M.; Coustau, C. The LBP/BPI multigenic family in invertebrates: Evolutionary history and evidences of specialization in mollusks. Dev. Comp. Immunol. 2016, 57, 20–30. [Google Scholar] [CrossRef]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; do Amaral, L.R.; et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 2017, 8, 15451. [Google Scholar] [CrossRef]
- Li, H.; Hambrook, J.R.; Pila, E.A.; Gharamah, A.A.; Fang, J.; Wu, X.; Hanington, P.C. Coordination of humoral immune factors dictates compatibility between Schistosoma mansoni and Biomphalaria glabrata. bioRxiv 2019, 767699. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.K.; Sreekumar, A.; Kundu, N.; Kathuria, R.; Verma, P.; Gandhi, S.; Chattopadhyay, K. Structural Basis and Functional Implications of the Membrane Pore-Formation Mechanisms of Bacterial Pore-Forming Toxins. Adv. Exp. Med. Biol. 2018, 1112, 281–291. [Google Scholar] [PubMed]
- Akiba, T.; Higuchi, K.; Mizuki, E.; Ekino, K.; Shin, T.; Ohba, M.; Kanai, R.; Harata, K. Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to β-pore-forming toxins. Proteins 2006, 63, 243–248. [Google Scholar] [CrossRef]
- Cole, A.R.; Gibert, M.; Popoff, M.; Moss, D.S.; Titball, R.W.; Basak, A.K. Clostridium perfringens ε-toxin shows structural similarity to the pore-forming toxin aerolysin. Nat. Struct. Mol. Biol. 2004, 11, 797–798. [Google Scholar] [CrossRef]
- Biomphalaria glabrata|VectorBase. Available online: https://www.vectorbase.org/organisms/biomphalaria-glabrata (accessed on 2 January 2020).
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The ConSurf Server. Available online: http://consurf.tau.ac.il/ (accessed on 2 January 2020).
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- InterPro. Available online: https://www.ebi.ac.uk/interpro/ (accessed on 2 January 2020).
- Crickmore, N.; Zeigler, D.R.; Feitelson, J.; Schnepf, E.; Rie, J.V.; Lereclus, D.; Baum, J.; Dean, D.H. Revision of the Nomenclature for the Bacillus thuringiensis Pesticidal Crystal Proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 807–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Deleury, E.; Dubreuil, G.; Elangovan, N.; Wajnberg, E.; Reichhart, J.-M.; Gourbal, B.; Duval, D.; Baron, O.L.; Gouzy, J.; Coustau, C. Specific versus Non-Specific Immune Responses in an Invertebrate Species Evidenced by a Comparative de novo Sequencing Study. PLoS ONE 2012, 7, e32512. [Google Scholar] [CrossRef] [PubMed]
- Portet, A.; Galinier, R.; Pinaud, S.; Portela, J.; Nowacki, F.; Gourbal, B.; Duval, D. BgTEP: An Antiprotease Involved in Innate Immune Sensing in Biomphalaria glabrata. Front. Immunol. 2018, 9, 1206. [Google Scholar] [CrossRef] [Green Version]
- Pinaud, S.; Portela, J.; Duval, D.; Nowacki, F.C.; Olive, M.-A.; Allienne, J.-F.; Galinier, R.; Dheilly, N.M.; Kieffer-Jaquinod, S.; Mitta, G.; et al. A Shift from Cellular to Humoral Responses Contributes to Innate Immune Memory in the Vector Snail Biomphalaria glabrata. PLoS Pathog. 2016, 12, e1005361. [Google Scholar] [CrossRef] [Green Version]
- Pinaud, S.; Portet, A.; Allienne, J.-F.; Belmudes, L.; Saint-Beat, C.; Arancibia, N.; Galinier, R.; Du Pasquier, L.; Duval, D.; Gourbal, B. Molecular characterisation of immunological memory following homologous or heterologous challenges in the schistosomiasis vector snail, Biomphalaria glabrata. Dev. Comp. Immunol. 2019, 92, 238–252. [Google Scholar] [CrossRef] [Green Version]
- Los, F.C.O.; Randis, T.M.; Aroian, R.V.; Ratner, A.J. Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol. Rev. 2013, 77, 173–207. [Google Scholar] [CrossRef] [Green Version]
- Popoff, M.R. Clostridial pore-forming toxins: powerful virulence factors. Anaerobe 2014, 30, 220–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, X.; Su, P.; Bi, D.; Tai, Z.; Li, Y.; Pang, Y.; Li, Q. Lamprey immune protein-1 (LIP-1) from Lampetra japonica induces cell cycle arrest and cell death in HeLa cells. Fish Shellfish Immunol. 2018, 75, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Feng, B.; Ren, Y.; Wu, D.; Chen, Y.; Huang, S.; Chen, S.; Xu, A. A pore-forming protein implements VLR-activated complement cytotoxicity in lamprey. Cell Discov. 2017, 3, 17033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, N.; Liu, N.; Cheng, W.; Jiang, Y.; Sun, H.; Chen, L.; Peng, J.; Zhang, Y.; Ding, Y.; Zhang, Z.; et al. Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein. EMBO Rep. 2016, 17, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Podobnik, M. Pore-forming toxins in Cnidaria. Dev. Biol. 2017, 72, 133–141. [Google Scholar] [CrossRef]
- Dang, L.; Rougé, P.; Van Damme, E.J.M. Amaranthin-Like Proteins with Aerolysin Domains in Plants. Front. Plant Sci. 2017, 8, 1368. [Google Scholar] [CrossRef] [Green Version]
- Manzano, S.; Megías, Z.; Martínez, C.; García, A.; Aguado, E.; Chileh, T.; López-Alonso, D.; García-Maroto, F.; Kejnovský, E.; Široký, J.; et al. Overexpression of a flower-specific aerolysin-like protein from the dioecious plant Rumex acetosa alters flower development and induces male sterility in transgenic tobacco. Plant J. 2017, 89, 58–72. [Google Scholar] [CrossRef] [Green Version]
- Hasan, I.; Gerdol, M.; Fujii, Y.; Rajia, S.; Koide, Y.; Yamamoto, D.; Kawsar, S.M.A.; Ozeki, Y. cDNA and Gene Structure of MytiLec-1, A Bacteriostatic R-Type Lectin from the Mediterranean Mussel (Mytilus galloprovincialis). Mar. Drugs 2016, 14, 92. [Google Scholar] [CrossRef] [Green Version]
- Jouiaei, M.; Sunagar, K.; Federman Gross, A.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an Ancient Venom: Recognition of a Novel Family of Cnidarian Toxins and the Common Evolutionary Origin of Sodium and Potassium Neurotoxins in Sea Anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef] [Green Version]
- Moran, Y.; Fredman, D.; Szczesny, P.; Grynberg, M.; Technau, U. Recurrent Horizontal Transfer of Bacterial Toxin Genes to Eukaryotes. Mol. Biol. Evol. 2012, 29, 2223–2230. [Google Scholar] [CrossRef] [Green Version]
- Szczesny, P.; Iacovache, I.; Muszewska, A.; Ginalski, K.; van der Goot, F.G.; Grynberg, M. Extending the Aerolysin Family: From Bacteria to Vertebrates. PLoS ONE 2011, 6, e20349. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. Why do we study animal toxins? Dongwuxue Yanjiu 2015, 36, 183–222. [Google Scholar]
- Chen, L.-L.; Xie, J.; Cao, D.-D.; Jia, N.; Li, Y.-J.; Sun, H.; Li, W.-F.; Hu, B.; Chen, Y.; Zhou, C.-Z. The pore-forming protein Aep1 is an innate immune molecule that prevents zebrafish from bacterial infection. Dev. Comp. Immunol. 2018, 82, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Yan, C.; Guo, X.; Zhou, K.; Li, S.; Gao, Q.; Wang, X.; Zhao, F.; Liu, J.; Lee, W.-H.; et al. Host-derived, pore-forming toxin–like protein and trefoil factor complex protects the host against microbial infection. Proc. Natl. Acad. Sci. USA 2014, 111, 6702–6707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sher, D.; Fishman, Y.; Zhang, M.; Lebendiker, M.; Gaathon, A.; Mancheño, J.-M.; Zlotkin, E. Hydralysins, a new category of beta-pore-forming toxins in cnidaria. J. Biol. Chem. 2005, 280, 22847–22855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sher, D.; Fishman, Y.; Melamed-Book, N.; Zhang, M.; Zlotkin, E. Osmotically driven prey disintegration in the gastrovascular cavity of the green hydra by a pore-forming protein. FASEB J. 2008, 22, 207–214. [Google Scholar] [CrossRef]
- Crisp, A.; Boschetti, C.; Perry, M.; Tunnacliffe, A.; Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 2015, 16, 50. [Google Scholar] [CrossRef] [Green Version]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Boto, L. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. Biol. Sci. 2014, 281, 20132450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Knapp, O.; Maier, E.; Benz, R.; Geny, B.; Popoff, M.R. Identification of the channel-forming domain of Clostridium perfringens Epsilon-toxin (ETX). Biochim. Biophys. Acta 2009, 1788, 2584–2593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiba, T.; Abe, Y.; Kitada, S.; Kusaka, Y.; Ito, A.; Ichimatsu, T.; Katayama, H.; Akao, T.; Higuchi, K.; Mizuki, E.; et al. Crystal Structure of the Parasporin-2 Bacillus thuringiensis Toxin That Recognizes Cancer Cells. J. Mol. Biol. 2009, 386, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Savva, C.G.; Clark, A.R.; Naylor, C.E.; Popoff, M.R.; Moss, D.S.; Basak, A.K.; Titball, R.W.; Bokori-Brown, M. The pore structure of Clostridium perfringens epsilon toxin. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Xu, C.; Wang, B.-C.; Yu, Z.; Sun, M. Structural Insights into Bacillus thuringiensis Cry, Cyt and Parasporin Toxins. Toxins (Basel) 2014, 6, 2732–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitada, S.; Abe, Y.; Maeda, T.; Shimada, H. Parasporin-2 requires GPI-anchored proteins for the efficient cytocidal action to human hepatoma cells. Toxicology 2009, 264, 80–88. [Google Scholar] [CrossRef]
- Okumura, S.; Saitoh, H.; Ishikawa, T.; Inouye, K.; Mizuki, E. Mode of action of parasporin-4, a cytocidal protein from Bacillus thuringiensis. Biochim. Biophys. Acta 2011, 1808, 1476–1482. [Google Scholar] [CrossRef] [Green Version]
- Ivie, S.E.; McClain, M.S. Identification of amino acids important for binding of Clostridium perfringens epsilon toxin to host cells and to HAVCR1. Biochemistry 2012, 51, 7588–7595. [Google Scholar] [CrossRef] [Green Version]
- Bokori-Brown, M.; Kokkinidou, M.C.; Savva, C.G.; Fernandes da Costa, S.; Naylor, C.E.; Cole, A.R.; Moss, D.S.; Basak, A.K.; Titball, R.W. Clostridium perfringens epsilon toxin H149A mutant as a platform for receptor binding studies. Protein Sci. 2013, 22, 650–659. [Google Scholar] [CrossRef] [Green Version]
- Aboul-Soud, M.A.M.; Al-Amri, M.Z.; Kumar, A.; Al-Sheikh, Y.A.; Ashour, A.E.; El-Kersh, T.A. Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells. Molecules 2019, 24, 506. [Google Scholar] [CrossRef] [Green Version]
- Chubicka, T.; Girija, D.; Deepa, K.; Salini, S.; Meera, N.; Raghavamenon, A.C.; Divya, M.K.; Babu, T.D. A parasporin from Bacillus thuringiensis native to Peninsular India induces apoptosis in cancer cells through intrinsic pathway. J. Biosci. 2018, 43, 407–416. [Google Scholar] [CrossRef]
- Brasseur, K.; Auger, P.; Asselin, E.; Parent, S.; Côté, J.-C.; Sirois, M. Parasporin-2 from a New Bacillus thuringiensis 4R2 Strain Induces Caspases Activation and Apoptosis in Human Cancer Cells. PLoS ONE 2015, 10, e0135106. [Google Scholar] [CrossRef]
- Saitoh, H.; Okumura, S.; Ishikawa, T.; Akao, T.; Mizuki, E.; Ohba, M. Investigation of a novel Bacillus thuringiensis gene encoding a parasporal protein, parasporin-4, that preferentially kills human leukemic T cells. Biosci. Biotechnol. Biochem. 2006, 70, 2935–2941. [Google Scholar] [CrossRef] [Green Version]
- Abe, Y.; Shimada, H.; Kitada, S. Raft-targeting and oligomerization of Parasporin-2, a bacillus thuringiensis crystal protein with anti-tumour activity. J. Biochem. 2008, 143, 269–275. [Google Scholar] [CrossRef]
- Ohba, M.; Nakashima, K.; Miyazaki, T. An optimum method for generation of functional recombinant apoptosis inhibitor of macrophage (AIM) protein. Seikagaku 2012, 84, 588–591. [Google Scholar]
- Chaisakul, J.; Hodgson, W.C.; Kuruppu, S.; Prasongsook, N. Effects of Animal Venoms and Toxins on Hallmarks of Cancer. J. Cancer 2016, 7, 1571–1578. [Google Scholar] [CrossRef]
- Tabata, A.; Ohkubo, Y.; Sakakura, E.; Tomoyasu, T.; Ohkura, K.; Nagamune, H. Investigation of a bacterial pore-forming chimera toxin for application as a novel drug-delivery system tool. Anticancer Res. 2012, 32, 2323–2329. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lassalle, D.; Tetreau, G.; Pinaud, S.; Galinier, R.; Crickmore, N.; Gourbal, B.; Duval, D. Glabralysins, Potential New β-Pore-Forming Toxin Family Members from the Schistosomiasis Vector Snail Biomphalaria glabrata. Genes 2020, 11, 65. https://doi.org/10.3390/genes11010065
Lassalle D, Tetreau G, Pinaud S, Galinier R, Crickmore N, Gourbal B, Duval D. Glabralysins, Potential New β-Pore-Forming Toxin Family Members from the Schistosomiasis Vector Snail Biomphalaria glabrata. Genes. 2020; 11(1):65. https://doi.org/10.3390/genes11010065
Chicago/Turabian StyleLassalle, Damien, Guillaume Tetreau, Silvain Pinaud, Richard Galinier, Neil Crickmore, Benjamin Gourbal, and David Duval. 2020. "Glabralysins, Potential New β-Pore-Forming Toxin Family Members from the Schistosomiasis Vector Snail Biomphalaria glabrata" Genes 11, no. 1: 65. https://doi.org/10.3390/genes11010065
APA StyleLassalle, D., Tetreau, G., Pinaud, S., Galinier, R., Crickmore, N., Gourbal, B., & Duval, D. (2020). Glabralysins, Potential New β-Pore-Forming Toxin Family Members from the Schistosomiasis Vector Snail Biomphalaria glabrata. Genes, 11(1), 65. https://doi.org/10.3390/genes11010065