Pathogenic Variant Filtering for Mitochondrial Genome Haplotype Reporting
Abstract
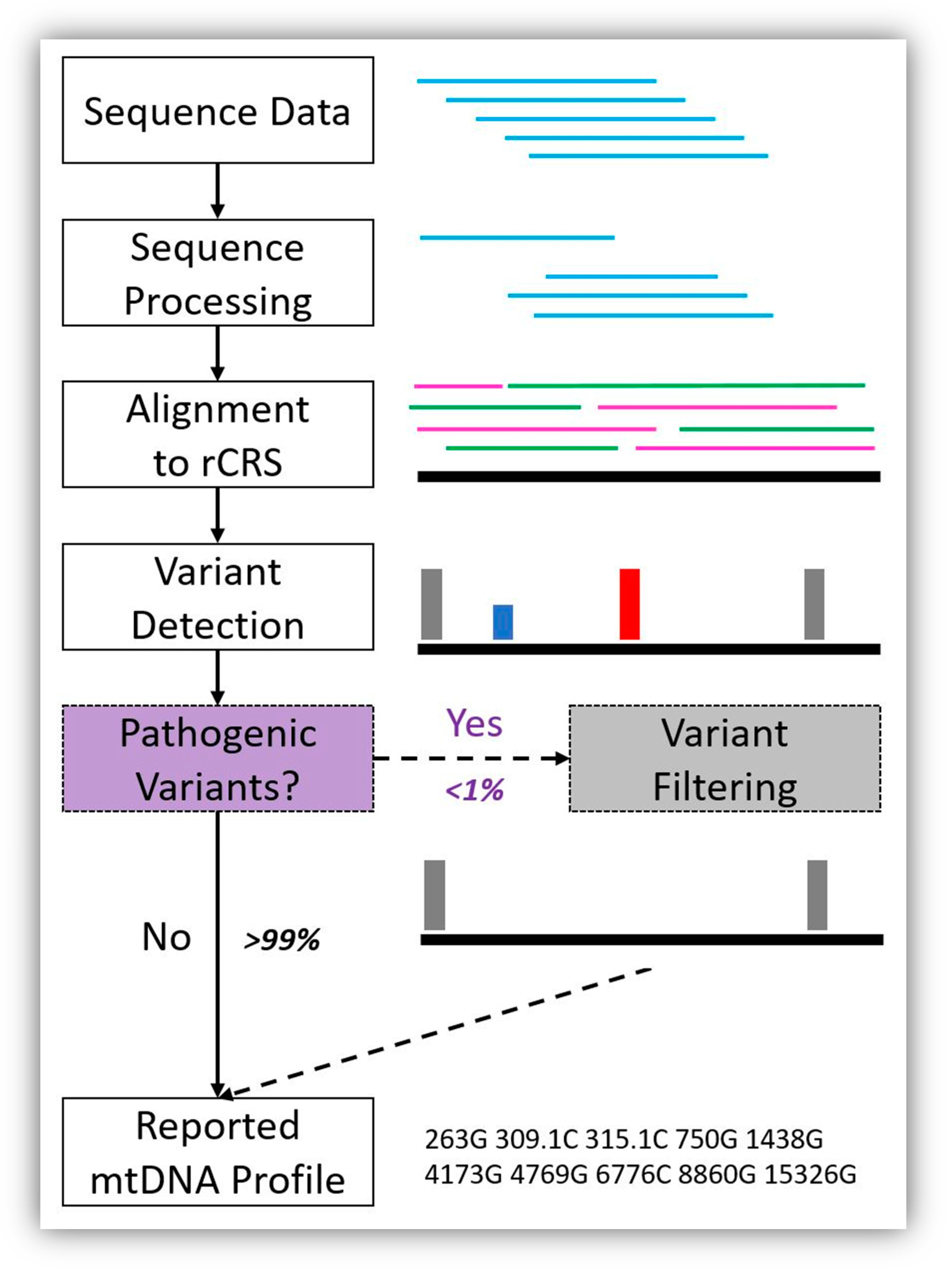
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- King, J.L.; LaRue, B.L.; Novroski, N.M.; Stoljarova, M.; Seo, S.B.; Zeng, X.; Warshauer, D.H.; Davis, C.P.; Parson, W.; Sajantila, A.; et al. High-Quality and High-Throughput Massively Parallel Sequencing of the Human Mitochondrial Genome using the Illumina MiSeq. Forensic Sci. Int. Genet. 2014, 12, 128–135. [Google Scholar] [CrossRef]
- Just, R.S.; Scheible, M.K.; Fast, S.A.; Sturk-Andreaggi, K.; Röck, A.W.; Bush, J.M.; Higginbotham, J.L.; Peck, M.A.; Ring, J.D.; Huber, G.E. Full mtGenome Reference Data: Development and Characterization of 588 Forensic-Quality Haplotypes Representing Three US Populations. Forensic Sci. Int. Genet. 2015, 14, 141–155. [Google Scholar] [CrossRef] [Green Version]
- García, Ó.; Alonso, S.; Huber, N.; Bodner, M.; Parson, W. Forensically Relevant Phylogeographic Evaluation of Mitogenome Variation in the Basque Country. Forensic Sci. Int. Genet. 2020, 46, 102260. [Google Scholar] [CrossRef]
- Parson, W.; Strobl, C.; Huber, G.; Zimmermann, B.; Gomes, S.M.; Souto, L.; Fendt, L.; Delport, R.; Langit, R.; Wootton, S. Evaluation of Next Generation mtGenome Sequencing using the Ion Torrent Personal Genome Machine (PGM). Forensic Sci. Int. Genet. 2013, 7, 543–549. [Google Scholar] [CrossRef] [Green Version]
- Marshall, C.; Sturk-Andreaggi, K.; Daniels-Higginbotham, J.; Oliver, R.S.; Barritt-Ross, S.; McMahon, T.P. Performance Evaluation of a Mitogenome Capture and Illumina Sequencing Protocol using Non-Probative, Case-Type Skeletal Samples: Implications for the use of a Positive Control in a Next-Generation Sequencing Procedure. Forensic Sci. Int. Genet. 2017, 31, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Strobl, C.; Eduardoff, M.; Bus, M.M.; Allen, M.; Parson, W. Evaluation of the Precision ID Whole MtDNA Genome Panel for Forensic Analyses. Forensic Sci. Int. Genet. 2018, 35, 21–25. [Google Scholar] [CrossRef]
- Eduardoff, M.; Xavier, C.; Strobl, C.; Casas-Vargas, A.; Parson, W. Optimized mtDNA Control Region Primer Extension Capture Analysis for Forensically Relevant Samples and Highly Compromised mtDNA of Different Age and Origin. Genes 2017, 8, 237. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Pesini, E.; Lott, M.T.; Procaccio, V.; Poole, J.C.; Brandon, M.C.; Mishmar, D.; Yi, C.; Kreuziger, J.; Baldi, P.; Wallace, D.C. An Enhanced MITOMAP with a Global mtDNA Mutational Phylogeny. Nucleic Acids Res. 2007, 35, D823–D828. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S. MITOMAP: Mitochondrial DNA Base Substitution Diseases: rRNA/tRNA Mutations with Cfrm Status. 2020. Available online: https://www.mitomap.org/foswiki/bin/view/MITOMAP/MutationsRNACfrm (accessed on 9 June 2020).
- Zhang, S. MITOMAP: Mitochondrial DNA Base Substitution Diseases: Coding and Control Region Point Mutations with Cfrm Status. 2020. Available online: https://www.mitomap.org/foswiki/bin/view/MITOMAP/MutationsCodingControlCfrm (accessed on 9 June 2020).
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and Revision of the Cambridge Reference Sequence for Human Mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [Green Version]
- Parson, W.; Dur, A. EMPOP—A Forensic mtDNA Database. Forensic Sci. Int. Genet. 2007, 1, 88–92. [Google Scholar] [CrossRef]
- Parson, W.; Gusmao, L.; Hares, D.; Irwin, J.; Mayr, W.; Morling, N.; Pokorak, E.; Prinz, M.; Salas, A.; Schneider, P. DNA Commission of the International Society for Forensic Genetics: Revised and Extended Guidelines for Mitochondrial DNA Typing. Forensic Sci. Int. Genet. 2014, 13, 134–142. [Google Scholar] [CrossRef]
- Parson, W.; Bandelt, H.J. Extended Guidelines for mtDNA Typing of Population Data in Forensic Science. Forensic Sci. Int. Genet. 2007, 1, 13–19. [Google Scholar] [CrossRef]
- Huber, N.; Parson, W.; Dür, A. Next Generation Database Search Algorithm for Forensic Mitogenome Analyses. Forensic Sci. Int. Genet. 2018, 37, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Guo, F.; Yu, J.; Liu, F.; Zhao, J.; Shen, H.; Zhao, B.; Jia, F.; Sun, Z.; Song, H. Strategies for Complete Mitochondrial Genome Sequencing on Ion Torrent PGM™ Platform in Forensic Sciences. Forensic Sci. Int. Genet. 2016, 22, 11–21. [Google Scholar] [CrossRef]
- Ma, K.; Zhao, X.; Li, H.; Cao, Y.; Li, W.; Ouyang, J.; Xie, L.; Liu, W. Massive Parallel Sequencing of Mitochondrial DNA Genomes from Mother-Child Pairs using the Ion Torrent Personal Genome Machine (PGM). Forensic Sci. Int. Genet. 2018, 32, 88–93. [Google Scholar] [CrossRef]
- Kutanan, W.; Kampuansai, J.; Srikummool, M.; Kangwanpong, D.; Ghirotto, S.; Brunelli, A.; Stoneking, M. Complete Mitochondrial Genomes of Thai and Lao Populations Indicate an Ancient Origin of Austroasiatic Groups and Demic Diffusion in the Spread of Tai-Kadai Languages. Hum. Genet. 2017, 136, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, Z.; He, G.; Wang, S.; Zou, X.; Liu, J.; Wang, F.; Ye, Z.; Hou, Y. Whole Mitochondrial Genome Analysis of Highland Tibetan Ethnicity using Massively Parallel Sequencing. Forensic Sci. Int. Genet. 2020, 44, 102197. [Google Scholar] [CrossRef]
- Mengge, W.; Guanglin, H.; Yongdong, S.; Shouyu, W.; Xing, Z.; Jing, L.; Zheng, W.; Hou, Y. Massively Parallel Sequencing of Mitogenome Sequences Reveals the Forensic Features and Maternal Diversity of Tai-Kadai-Speaking Hlai Islanders. Forensic Sci. Int. Genet. 2020, 47, 102303. [Google Scholar] [CrossRef]
- Ramos, A.; Santos, C.; Mateiu, L.; Gonzalez Mdel, M.; Alvarez, L.; Azevedo, L.; Amorim, A.; Aluja, M.P. Frequency and Pattern of Heteroplasmy in the Complete Human Mitochondrial Genome. PLoS ONE 2013, 8, e74636. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.; Strobl, C.; Vullo, C.; Catelli, L.; Machado, P.; Huber, N.; Schnaller, L.; Huber, G.; Xavier, C.; Carvalho, E.F. The Maternal Inheritance of Alto Paraná Revealed by Full Mitogenome Sequences. Forensic Sci. Int. Genet. 2019, 39, 66–72. [Google Scholar] [CrossRef]
- Avila, E.; Graebin, P.; Chemale, G.; Freitas, J.; Kahmann, A.; Alho, C. Full mtDNA Genome Sequencing of Brazilian Admixed Populations: A Forensic-Focused Evaluation of a MPS Application as an Alternative to Sanger Sequencing Methods. Forensic Sci. Int. Genet. 2019, 42, 154–164. [Google Scholar] [CrossRef]
- Taylor, C.; Kiesler, K.; Sturk-Andreaggi, K.; Ring, J.D.; Parson, W.; Vallone, P.; Marshall, C. Platinum Quality Mitogenome Haplotypes form U.S. Populations. Unpublished work.
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and Organization of the Human Mitochondrial Genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- van Oven, M. PhyloTree Build 17: Growing the Human Mitochondrial DNA Tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e392–e394. [Google Scholar] [CrossRef] [Green Version]
- Sturk-Andreaggi, K.; Peck, M.A.; Boysen, C.; Dekker, P.; McMahon, T.P.; Marshall, C.K. AQME: A Forensic Mitochondrial DNA Analysis Tool for Next-Generation Sequencing Data. Forensic Sci. Int. Genet. 2017, 31, 189–197. [Google Scholar] [CrossRef]
- Holland, M.M.; Pack, E.D.; McElhoe, J.A. Evaluation of GeneMarker® HTS for Improved Alignment of mtDNA MPS Data, Haplotype Determination, and Heteroplasmy Assessment. Forensic Sci. Int. Genet. 2017, 28, 90–98. [Google Scholar] [CrossRef]
- Budowle, B.; Gyllensten, U.; Chakraborty, R.; Allen, M. Forensic analysis of the mitochondrial coding region and association to disease. Int. J. Legal Med. 2005, 119, 314–315. [Google Scholar] [CrossRef]

| Homoplasmic Variants with Confirmed Pathogenicity | Heteroplasmic Variants with Confirmed Pathogenicity |
|---|---|
| T616C, C1494T, A1555G, C3303T, G3376A, G3460A, G3635A, G3697A, G3700A, G3733A, C4171A, A4300G, A7445G, C7471CC, G7497A, T7511C, T8528C, T8851C, T8993G, T9035C, T9176C, T9176G, T9185C, T9205del A9206del, T10158C, G10197A, T10663C, G11778A, G13051A, T13094C, G14459A, C14482A, C14482G, T14484C, C14568T, T14674C, T14709C | G583A, T616C, G1606A, A1630G, G1644A, A3243G, A3243T, C3256T, T3258C, A3260G, T3271del, T3271C, A3280G, T3291C, A3302G, C3303T, G3376A, G3460A, G3697A, G3733A, G3890A, A3902G C3904A T3905A T3906G C3908T, C4171A, G4298A, A4300G, G4308A, G4332A, G4450A, G5521A, A5537AT, G5650A, A5690G, G5703A, T5728C, A7445G, C7471CC, G7497A, T7510C, T7511C, T8306C, G8313A, G8340A, A8344G, T8356C, G8363A, T8528C, T8851C, G8969A, T8993C, T8993G, T9035C, A9155G, T9176C, T9176G, T9185C, T10010C, T10158C, T10191C, G10197A, C11777A, G11778A, G12147A, C12258A, G12276A, G12294A, G12315A, G12316A, T12706C, G13042A, T13094C, G13513A, A13514G, G14459A, C14482A, C14482G, T14484C, T14487C, A14495G, T14709C, G14710A, T14849C, T14864C, A15579G |
| Count of Observations | Observed Frequency (n = 26,013) | Confirmed Pathogenic Variants Observed |
|---|---|---|
| 1 | 0.004% | T616C, C4171A, A4300G, G5703A(R), A7445G, T9176G, T9185C(Y), T10663C, G12276A(R), G13051A, G13513A(R), G14459A(R), T14484C(Y), T14709C(Y) |
| 2 | 0.008% | G3460A(R), G3700A, G3733A, C4171A(M), T8851C, G11778A(R), C14482A |
| 3 | 0.012% | C1494T, T8993G, T9176C, T9185C, G10197A, G14459A |
| 4 | 0.015% | G3635A, C7471CC, T14674C |
| 6 | 0.023% | C14568T |
| 25 | 0.096% | G3460A |
| 37 | 0.142% | A1555G |
| 55 | 0.211% | T14484C |
| 131 | 0.504% | G11778A |
| Original Dataset | Filtered Dataset | |||||||
|---|---|---|---|---|---|---|---|---|
| Population | Samples | Unique Haplotypes | RMP | Haplotype Diversity | Pathogenic Variants Filtered | Change in Unique Haplotypes | Change in RMP | Change in Haplotype Diversity |
| West Eurasian | 623 | 575 (30 shared) | 0.21% | 0.9995 | 1 | 0 | 0% | 0 |
| African | 613 | 597 (15 shared) | 0.17% | 0.9999 | 3 | 0 | 0% | 0 |
| East Asian | 630 | 557 (45 shared) | 0.22% | 0.9994 | 4 | 0 | 0% | 0 |
| Hispanic/Native American | 622 | 568 (43 shared) | 0.20% | 0.9996 | 1 | 0 | 0% | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marshall, C.; Sturk-Andreaggi, K.; Ring, J.D.; Dür, A.; Parson, W. Pathogenic Variant Filtering for Mitochondrial Genome Haplotype Reporting. Genes 2020, 11, 1140. https://doi.org/10.3390/genes11101140
Marshall C, Sturk-Andreaggi K, Ring JD, Dür A, Parson W. Pathogenic Variant Filtering for Mitochondrial Genome Haplotype Reporting. Genes. 2020; 11(10):1140. https://doi.org/10.3390/genes11101140
Chicago/Turabian StyleMarshall, Charla, Kimberly Sturk-Andreaggi, Joseph D. Ring, Arne Dür, and Walther Parson. 2020. "Pathogenic Variant Filtering for Mitochondrial Genome Haplotype Reporting" Genes 11, no. 10: 1140. https://doi.org/10.3390/genes11101140
APA StyleMarshall, C., Sturk-Andreaggi, K., Ring, J. D., Dür, A., & Parson, W. (2020). Pathogenic Variant Filtering for Mitochondrial Genome Haplotype Reporting. Genes, 11(10), 1140. https://doi.org/10.3390/genes11101140






