Genome-Wide Analysis of Chemosensory Protein Genes (CSPs) Family in Fig Wasps (Hymenoptera, Chalcidoidea)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequence Sources
2.2. Manual Annotation and Identification of CSPs
2.3. Phylogenetic Analyses
2.4. Analysis of CSPs Characteristics
2.5. CSP Gene Family Expansion and Contraction
2.6. Gene Expression Pattern Predicted by Codon Adaptation Index Analysis
3. Results
3.1. Identification of CSPs Genes in the 11 Fig Wasp Species
3.2. Characterization of the CSPs Genes
3.3. Multiple Sequence Alignment, Phylogenetic Analysis and Classification of the CSP Genes
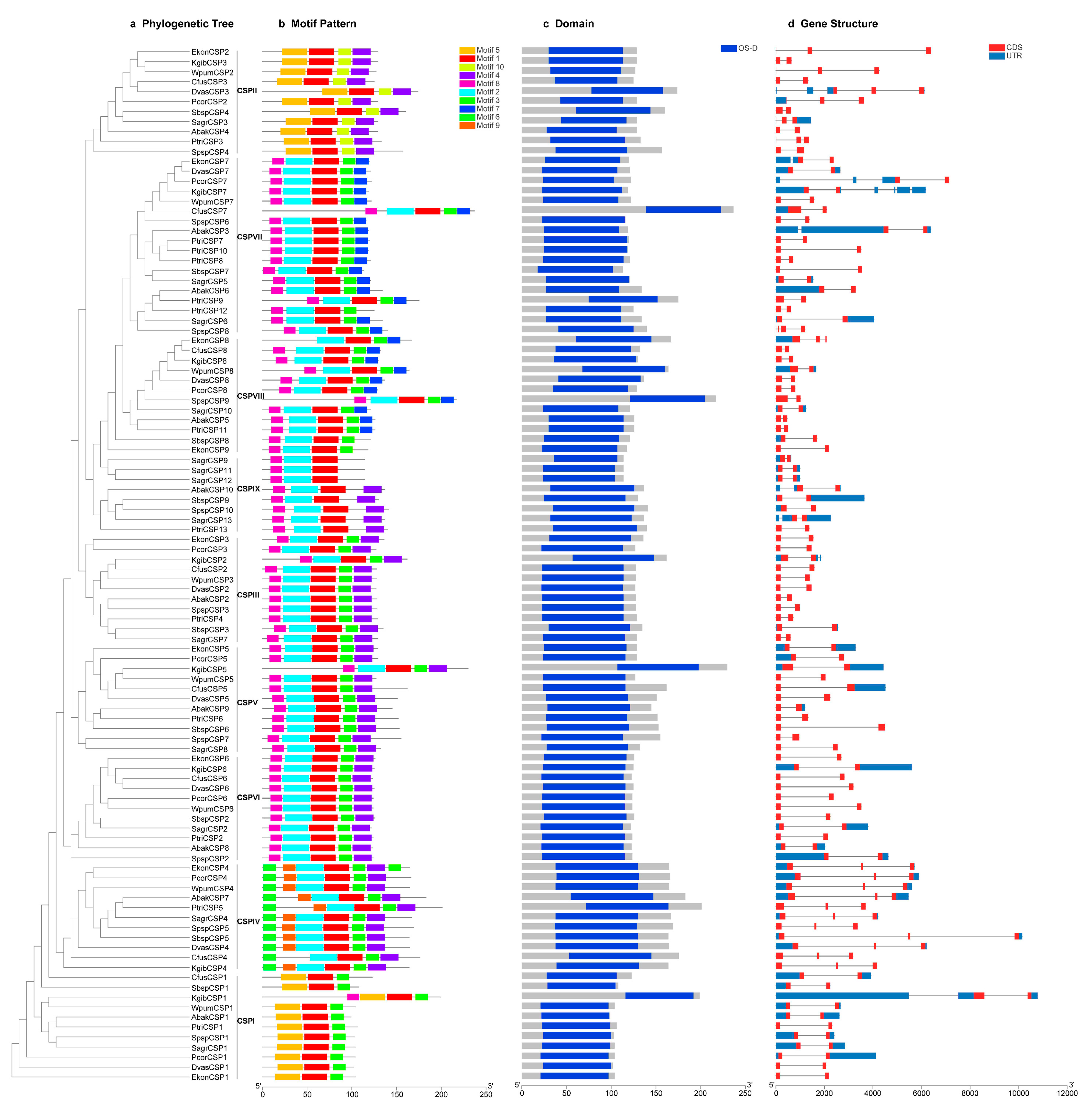
3.4. Comprehensive Analyses of the Gene Tree, Conserved Motifs, Domains and Gene Structures of the CSP Genes
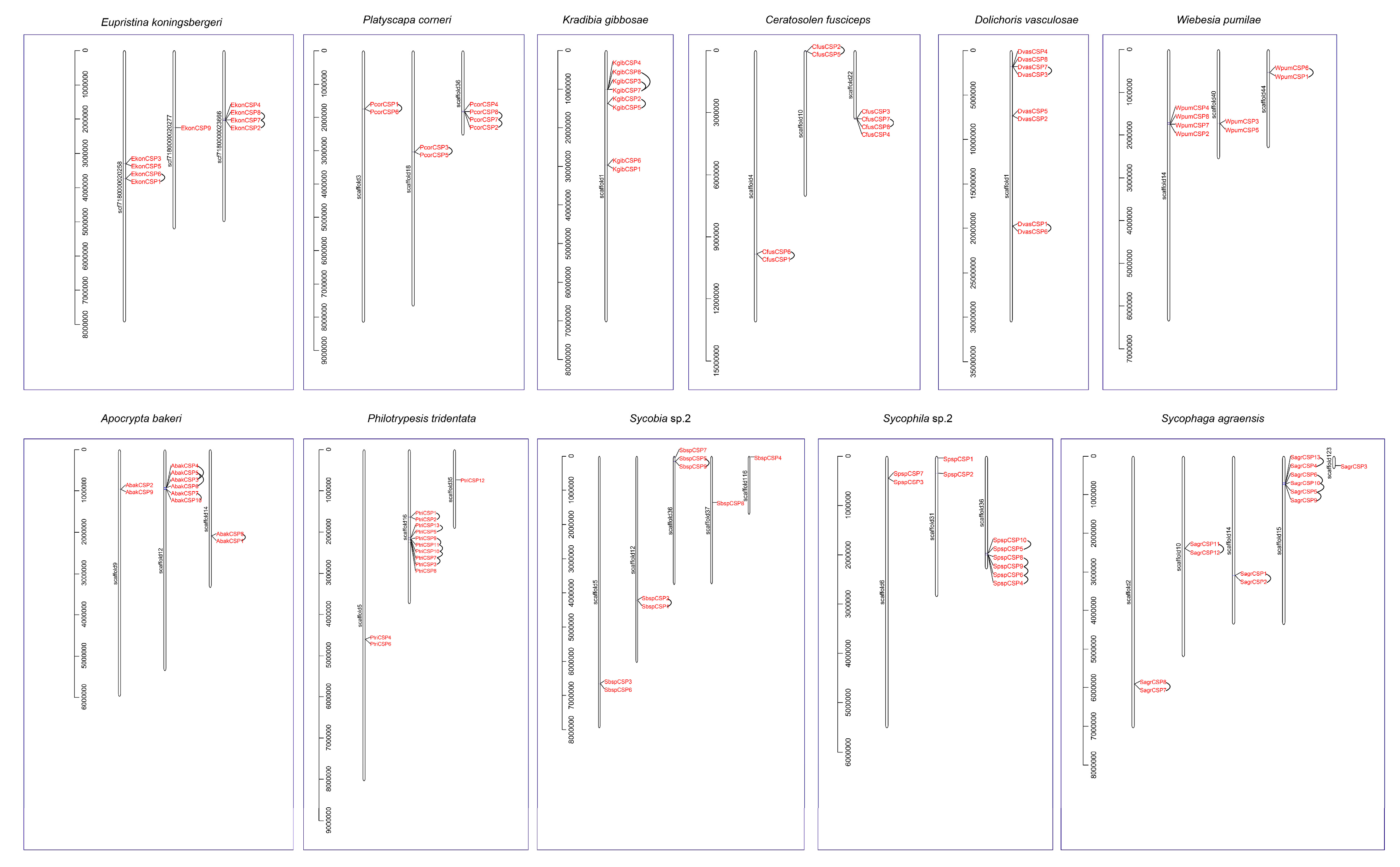
3.5. Distribution and Tandem Analysis of CSP Genes on the Genomic Scaffolds
3.6. CSPs Gene Family Expansion and Contraction
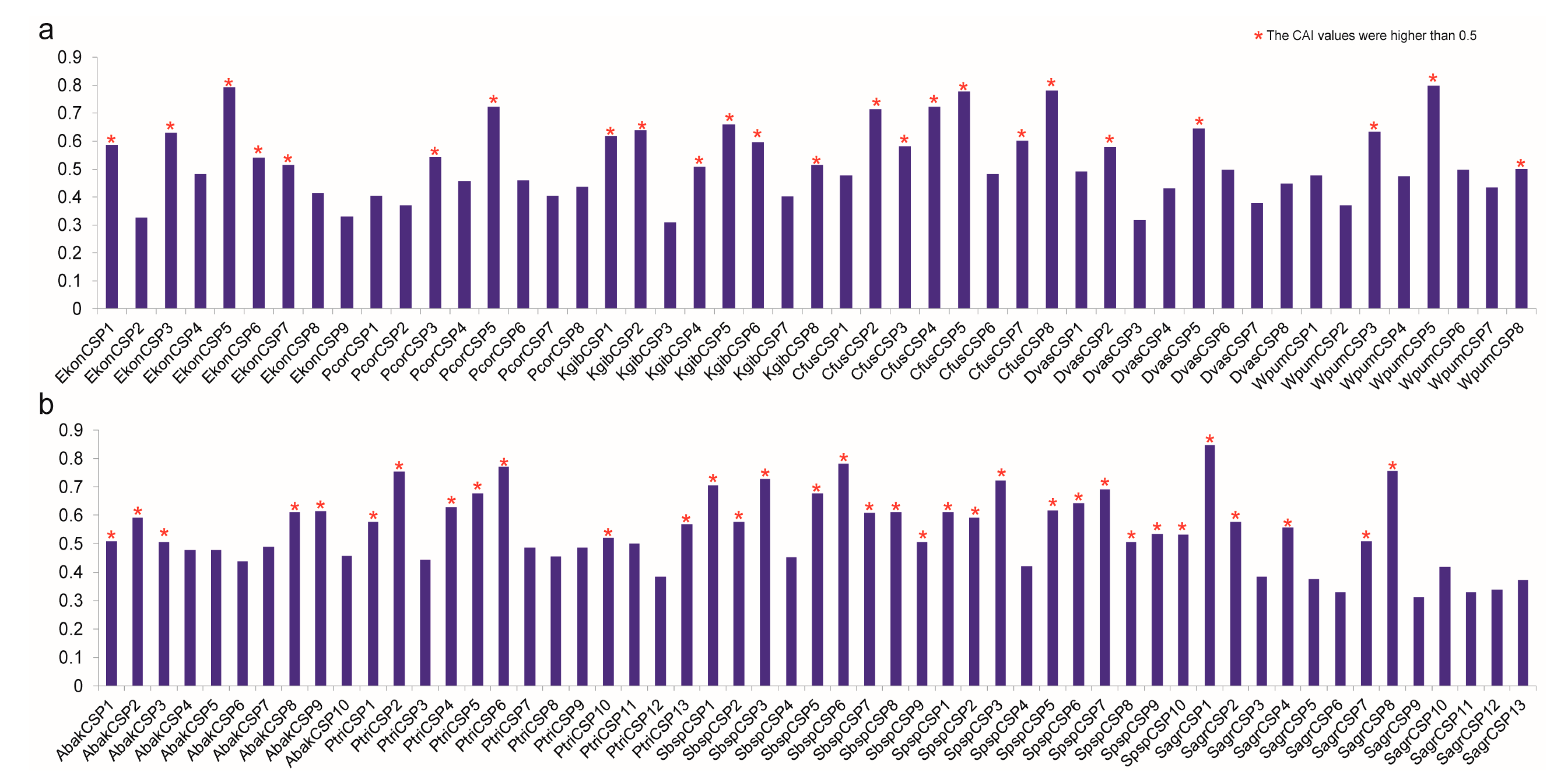
3.7. Codon Adaptation Index Analysis of CSPs to Predict their Expression Indirectly
4. Discussion
4.1. The CSP Gene Family is Conserved across the Genomes of Fig Wasps
4.2. The CSP Gene Family Is Streamlined in the Pollinating Fig Wasps
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smadja, C.; Butlin, R.K. On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity 2009, 102, 77–97. [Google Scholar] [CrossRef] [Green Version]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Eyun, S.; Soh, H.Y.; Posavi, M.; Munro, J.B.; Hughes, D.S.T.; Murali, S.C.; Qu, J.X.; Dugan, S.; Lee, S.L.; Chao, H.; et al. Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol. Biol. Evol. 2017, 34, 1838–1862. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Snyder, S.H.; Sklar, P.B.; Pevsner, J. Molecular mechanisms of olfaction. J. Biol. Chem. 1988, 263, 13971–13974. [Google Scholar] [CrossRef]
- Rützler, M.; Zwiebel, L.J. Molecular biology of insect olfaction: Recent progress and conceptual models. J. Comp. Physiol. A 2005, 191, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Kaupp, U.B. Olfactory signalling in vertebrates and insects: Differences and commonalities. Nat. Rev. Neurosci. 2010, 11, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Maida, R. Odorant-binding proteins in vertebrates and insects: Similarities and possible common function. Chem. Senses 1990, 15, 205–215. [Google Scholar] [CrossRef]
- Pelosi, P.; Maida, R. Odorant-binding proteins in insects. Comp. Biochem. Phys. B 1995, 111, 503–514. [Google Scholar] [CrossRef]
- Smith, D.P. Odor and pheromone detection in Drosophila melanogaster. Pflug. Arch. Eur. J. Phy. 2007, 454, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.R.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biological Reviews of the Cambridge Philosophical Society 2018, 93, 184–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaissling, K.E. Olfactory perireceptor and receptor events in moths: A kinetic model. Chem. Senses 2001, 26, 125–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, W.S.; Chen, A.M.; Ishida, Y.; Chiang, V.P.; Erickson, M.L.; Morgan, T.I.; Tsuruda, J.M. Kinetics and molecular properties of pheromone binding and release. P. Natl. Acad. Sci. USA 2005, 102, 5386–5391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.J.; Vieira, F.G.; He, X.; Smadja, C.M.; Liu, R.; Rozas, J.; Field, L.M. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Dani, F.R.; Michelucci, E.; Francese, S.; Mastrobuoni, G.; Cappellozza, S.; Marca, G.L.; Niccolini, A.; Felicioli, A.; Moneti, G.; Pelosi, P. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem. Senses 2011, 36, 335–344. [Google Scholar] [CrossRef]
- Pelosi, P.; Calvello, M.; Ban, L.P. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 2005, 30, i291–i292. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar]
- Wanner, K.W.; Willis, L.G.; Theilmann, D.A.; Isman, M.B.; Feng, Q.L.; Plettner, E. Analysis of the insect os-d-like gene family. J. Chem. Ecol. 2004, 30, 889–911. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [Green Version]
- Lartigue, A.; Campanacci, V.; Roussel, A.; Larsson, A.M.; Jones, T.A.; Tegoni, M.; Cambillau, C. X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 2002, 277, 32094–32098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.N.; Ye, Z.F.; Yang, K.; Dong, S.L. Antenna-predominant and male-biased CSP19 of Sesamia inferens is able to bind the female sex pheromones and host plant volatiles. Gene 2014, 536, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.H.; Yue, Z.; Jia, L.Y.; Yang, X.H.; Niu, L.H.; Wang, Z.; Zhang, P.; Sun, B.F.; He, S.M.; Li, Z.; et al. Obligate mutualism within a host drives the extreme specialization of a fig wasp genome. Genome Biol. 2013, 14, R141. [Google Scholar] [CrossRef] [Green Version]
- Cook, J.M. Alternative mating tactics and fatal fighting in male fig wasps. Insect Evolutionary Ecology 2005, 83–110. [Google Scholar]
- Werren, J.H.; Richards, S.; Desjardins, C.A.; Niehuis, O.; Gadau, J.; Colbourne, J.K.; Beukeboom, L.W.; Desplan, C.; Elsik, C.G.; Grimmelikhuijzen, C.J.P.; et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 2010, 327, 343–348. [Google Scholar] [CrossRef]
- Forêt, S.; Wanner, K.W.; Maleszka, R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Molec. 2007, 37, 19–28. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. A graphical viewer of phylogenetic trees. In FigTree v1.4.0.; FigTree: Edinburgh, UK, 2012. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [Green Version]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct 2008, 3, 38. [Google Scholar]
- Xu, Y.L.; He, P.; Zhang, L.; Fang, S.Q.; Dong, S.L.; Zhang, Y.J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics 2009, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Sharp, P.M.; Li, W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [Green Version]
- Pandey, A.; Suman, S.; Chandna, S. Predictive role of mitochondrial genome in the stress resistance of insects and nematodes. Bioinformation 2010, 5, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Khandia, R.; Singhal, S.; Kumar, U.; Ansari, A.; Tiwari, R.; Dhama, K.; Das, J.; Munjal, A.; Singh, R.K. Analysis of nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019, 10, 886. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.P.; Feng, Y.; Xue, Q.Z. Analysis of factors shaping codon usage in the mitochondrion genome of Oryza sativa. Mitochondrion 2004, 4, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.; Mukherjee, I.; Choudhury, M.; Das, S. Unusual codon usage bias in low expression genes of Vibrio cholerae. Bioinformation 2008, 3, 213–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.H. An improved implementation of codon adaptation index. Evol. Bioinform. 2007, 3, 53–58. [Google Scholar] [CrossRef]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.L. Pherokine-2 and-3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 2003, 270, 3398–3407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wu, N.N.; Song, W.L.; Yin, G.J.; Qin, Y.J.; Yan, Y.M.; Hu, Y.K. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Zhang, L.Q. Tandemly arrayed genes in vertebrate genomes. Comp. Funct. Genom. 2008, 2008, 545269. [Google Scholar] [CrossRef] [Green Version]
- Reams, A.B.; Neidle, E.L. Selection for gene clustering by tandem duplication. Annu. Rev. Microbiol. 2004, 58, 119–142. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Kondrashov, F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. P. Roy. Soc. B Biol. Sci. 2012, 279, 5048–5057. [Google Scholar] [CrossRef] [Green Version]
- Peters, R.S.; Niehuis, O.; Gunkel, S.; Bläser, M.; Mayer, C.; Podsiadlowski, L.; Kozlov, A.; Donath, A.; van Noort, S.; Liu, S.; et al. Transcriptome sequence-based phylogeny of chalcidoid wasps (Hymenoptera: Chalcidoidea) reveals a history of rapid radiations, convergence, and evolutionary success. Mol. Phylogenet. Evol. 2018, 120, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, T.; Fu, W.B.; Li, B.; He, Z.B.; Chen, B. Comparative genomics of chemosensory protein genes (CSPs) in twenty-two mosquito species (Diptera: Culicidae): Identification, characterization, and evolution. PLoS ONE 2018, 13, e0190412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Zhou, X.; Zhang, C.X.; Yu, L.L.; Fan, H.W.; Wang, Z.; Xu, H.J.; Xi, Y.; Zhu, Z.R.; Zhou, W.W.; et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014, 15, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dippel, S.; Oberhofer, G.; Kahnt, J.; Gerischer, L.; Opitz, L.; Schachtner, J.; Stanke, M.; Schutz, S.; Wimmer, E.A.; Angeli, S. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 2014, 15, 1141. [Google Scholar] [CrossRef] [Green Version]
- You, M.S.; Yue, Z.; He, W.Y.; Yang, X.H.; Yang, G.; Xie, M.; Zhan, D.L.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Lin, Y.; Xia, Q.Y.; Xiang, Z.H. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem. Molec. 2007, 37, 266–277. [Google Scholar] [CrossRef]
- Zhou, X.H.; Ban, L.P.; Iovinella, I.; Zhao, L.J.; Gao, Q.; Felicioli, A.; Sagona, S.; Pieraccini, G.; Pelosi, P.; Zhang, L.; et al. Diversity, abundance, and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis. Biol. Chem. 2013, 394, 43–54. [Google Scholar] [CrossRef]
- Kirkness, E.F.; Haas, B.J.; Sun, W.; Braig, H.R.; Perotti, M.A.; Clark, J.M.; Lee, S.H.; Robertson, H.M.; Kennedy, R.C.; Elhaik, E.; et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. P. Natl. Acad. Sci. USA 2010, 107, 12168–12173. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Wang, N.X.; Niu, L.M.; Bian, S.N.; Xiao, J.H.; Huang, D.W. Odorant-binding protein (OBP) genes affect host specificity in a fig-pollinator mutualistic system. Insect Mol. Biol. 2014, 23, 621–631. [Google Scholar] [CrossRef]
- Matsuo, T. Genes for host-plant selection in Drosophila. J. Neurogenet. 2008, 22, 195–210. [Google Scholar] [CrossRef] [PubMed]



| Species | CSP Gene Number |
|---|---|
| Eupristina koningsbergeri | 9 |
| Platyscapa corneri | 8 |
| Kradibia gibbosae | 8 |
| Ceratosolen fusciceps | 8 |
| Dolichoris vasculosae | 8 |
| Wiebesia pumilae | 8 |
| Apocrypta bakeri | 10 |
| Philotrypesis tridentata | 13 |
| Sycobia sp.2 | 9 |
| Sycophila sp.2 | 10 |
| Sycophaga agraensis | 13 |
| Ceratosolen solms | 7 |
| Nasonia vitripennis | 9 |
| Apis mellifera | 6 |
| Drosophila melanogaster | 4 |
| Solenopsis invicta | 21 |
| Culex quinquefasciatus | 27 |
| Aedes albopictus | 83 |
| Nilaparvata lugens | 17 |
| Tribolium castaneum | 20 |
| Plutella xylostella | 32 |
| Bombyx mori | 20 |
| Locusta migratoria | 70 |
| Pediculus humanus | 7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Z.; Huang, D.; Zhao, D.; Li, J.; Wei, X.; Xiao, J. Genome-Wide Analysis of Chemosensory Protein Genes (CSPs) Family in Fig Wasps (Hymenoptera, Chalcidoidea). Genes 2020, 11, 1149. https://doi.org/10.3390/genes11101149
Xin Z, Huang D, Zhao D, Li J, Wei X, Xiao J. Genome-Wide Analysis of Chemosensory Protein Genes (CSPs) Family in Fig Wasps (Hymenoptera, Chalcidoidea). Genes. 2020; 11(10):1149. https://doi.org/10.3390/genes11101149
Chicago/Turabian StyleXin, Zhaozhe, Dawei Huang, Dan Zhao, Jiaxing Li, Xianqin Wei, and Jinhua Xiao. 2020. "Genome-Wide Analysis of Chemosensory Protein Genes (CSPs) Family in Fig Wasps (Hymenoptera, Chalcidoidea)" Genes 11, no. 10: 1149. https://doi.org/10.3390/genes11101149




