Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis
Abstract
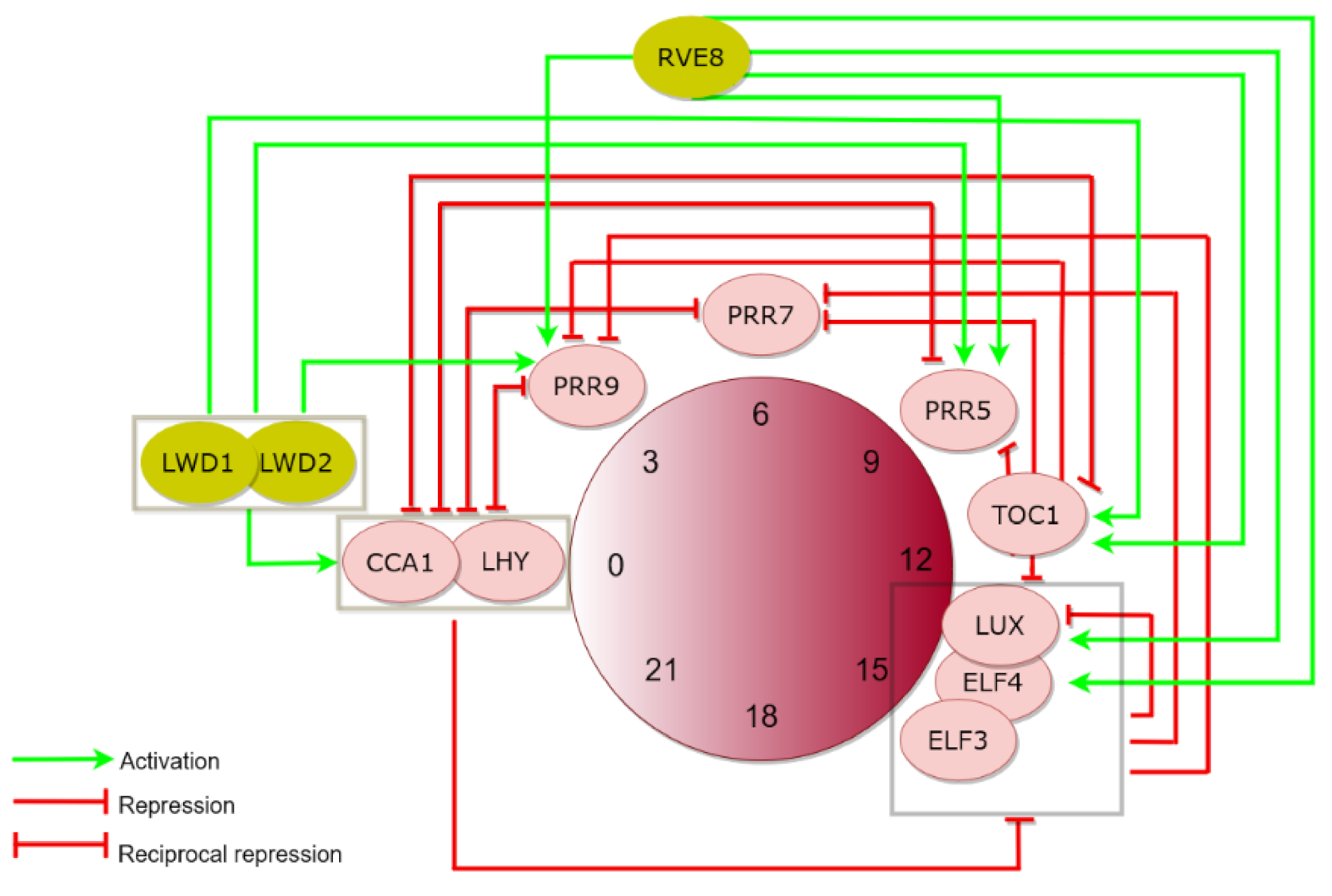
:1. The Plant Circadian Clock
2. Transcriptional Dynamics and Chromatin Status
3. Functional Link between Chromatin and Circadian Transcription
4. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gachon, F.; Nagoshi, E.; Brown, S.A.; Ripperger, J.; Schibler, U. The mammalian circadian timing sytem: From gene expression to physiology. Chromosoma 2004, 113, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.A. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol. Metab. 2016, 27, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.H.; Takahashi, J.S. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, C.J.; Kay, S.A. Circadian surprise—It’s not all about transcription. Science 2012, 338, 338–400. [Google Scholar] [CrossRef] [PubMed]
- Young, M.W.; Kay, S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Gen. 2001, 2, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tingay, S.; Wang, Z.Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resco, V.; Hartwell, J.; Hall, A. Ecological implications of plants ability to tell the time. Ecol. Lett. 2009, 12, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, S.; Green, R.M. Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 2009, 12, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef] [PubMed]
- Más, P. Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008, 18, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Mas, P. Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell. 2014, 26, 79–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateos, J.L.; de Leone, M.J.; Torchio, J.; Reichel, M.; Staiger, D. Beyond transcription: Fine-tuning of circadian timekeeping by post-transcriptional regulation. Genes 2018, 9, 616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Wang, J.; Huang, F.Y.; Yang, S.; Wu, K. The plant circadian clock and chromatin modifications. Genes 2018, 9, 561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef]
- Millar, A.J.; Carré, I.A.; Strayer, C.A.; Chua, N.H.; Kay, S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 1995, 267, 1161–1163. [Google Scholar] [CrossRef]
- DSomers, E.; Webb, A.A.R.; Pearson, M.; Kay, S.A. The short-period mutant toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998, 125, 485–494. [Google Scholar]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Mas, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [Green Version]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Más, P.; Alabadí, D.; Yanovsky, M.J.; Oyama, T.; Kay, S.A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 2003, 15, 223–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Nakamichi, N. Pseudo-Response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 2005, 46, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Makino, S.; Kiba, T.; Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, S.; Manfield, I.; Stockley, P.; Carré, I.A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 2015, 10, e0143943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, K.A.; Albertson, T.M.; Wagner, D.R. EARLY FLOWERING3 Encodes a Novel Protein That Regulates Circadian Clock Function and Flowering in Arabidopsis. Plant Cell 2001, 13, 1281–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognár, L.; Nagy, F.; Millar, A.J. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef] [Green Version]
- Onai, K.; Ishiura, M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005, 10, 963–972. [Google Scholar] [CrossRef]
- McWatters, H.G.; Kolmos, E.; Hall, A.; Doyle, M.R.; Amasino, R.M.; Gyula, P.; Nagy, F.; Millar, A.J.; Davis, S.J. ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 2007, 144, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Kolmos, E.; Nowak, M.; Werner, M.; Fischer, K.; Schwarz, G.; Mathews, S.; Schoof, H.; Nagy, F.; Bujnicki, J.M.; Davis, S.J. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009, 3, 350–366. [Google Scholar] [CrossRef] [Green Version]
- McWatters, H.G.; Bastow, R.M.; Hall, A.; Millar, A.J. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 2000, 408, 716–720. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezer, D.; Jung, J.H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stockle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 Regulates GIGANTEA Chromatin Access through Subnuclear Sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto, C.; López-Salmerón, V.; Davière, J.-M.; Prat, S. ELF3-PIF4 Interaction Regulates Plant Growth Independently of the Evening Complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyam, T.; Kinoshita, T.; Nakamichi, N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef] [Green Version]
- Pokhilko, A.; Fernandez, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1,is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 335, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Portolés, S.; Más, P. The functional interplay between protein kinase CK2 and cca1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010, 6, e1001201. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Siddiqui, H.; Teng, Y.; Lin, R.; Wan, X.; Li, J.; Lau, O.-S.; Ouyang, X.; Dai, M.; Wan, J.; et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 2011, 13, 616–622. [Google Scholar] [CrossRef]
- SLu, X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.J.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 Interact in the Control of Hypocotyl Length and Flowering Time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar]
- Dixon, L.E.; Knox, K.; Kozma-Bognar, L.; Southern, M.M.; Pokhilko, A.; Millar, A.J. Temporal Repression of Core Circadian Genes Is Mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011, 21, 120–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, B.Y.; Helfer, A.; Nusinow, D.A.; Kay, S.A. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav. 2012, 7, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.F.; Wang, Y.; Wu, S.H. Two new clock proteins, LWD1 and LWD2, regulate arabidopsis photoperiodic flowering. Plant Physiol. 2008, 148, 948–959. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wu, J.F.; Nakamichi, N.; Sakakibara, H.; Nam, H.G.; Wu, S.H. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 Form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011, 23, 486–498. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef] [Green Version]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef]
- Xie, Q.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.; Zhang, C.; Li, Y.; Xing, H.; Zhi, L.; Yue, Z.; et al. LNK1 and LNK2 Are Transcriptional Coactivators in the Arabidopsis Circadian Oscillator. Plant Cell Online 2014, 26, 2843–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Mas, P. Interactive roles of chromatin regulation and circadian clock function in plants. Genome Biol. 2019, 20, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, S.J.; Ronald, J. Making the clock tick: The transcriptional landscape of the plant circadian clock. F1000Research 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Pfluger, J.; Wagner, D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007, 10, 645–652. [Google Scholar] [CrossRef] [Green Version]
- Pikaard, C.S.; Scheid, O.M. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Probst, A.V.; Desvoyes, B.; Gutierrez, C. Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 2020, 71, 5191–5204. [Google Scholar] [CrossRef]
- Dong, P.; Tu, X.; Liang, Z.; Kang, B.-H.; Zhong, S. Plant and animal chromatin three-dimensional organization: Similar structures but different functions. J. Exp. Bot. 2020, 71, 5119–5128. [Google Scholar] [CrossRef]
- Chen, Z.J.; Tian, L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 2007, 1769, 295–307. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Servet, C.; Silva, N.C.E.; Zhou, D.X. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Singh, R.; Lodhi, N. Histone demethylases and control of gene expression in plants. Cell. Mol. Biol. 2014, 60, 97–105. [Google Scholar]
- Perales, M.; Más, P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 2007, 19, 2111–2123. [Google Scholar] [CrossRef] [Green Version]
- Song, H.R.; Noh, Y.S. Rhythmic Oscillation of Histone Acetylation and Methylation at the Arabidopsis Central Clock Loci. Mol. Cells 2012, 34, 279–287. [Google Scholar] [CrossRef]
- Malapeira, J.; Khaitova, L.C.; Mas, P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2012, 109, 21540–21545. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Seo, P.J. The HAF2 protein shapes histone acetylation levels of PRR5 and LUX loci in Arabidopsis. Planta 2018, 248, 513–518. [Google Scholar] [CrossRef]
- Lee, H.G.; Hong, C.; Seo, P.J. The Arabidopsis sin3-hdac complex facilitates temporal histone deacetylation at the Cca1 and Prr9 loci for robust circadian oscillation. Front. Plant Sci. 2019, 10, 171. [Google Scholar] [CrossRef]
- Lee, K.; Mas, P.; Seo, P.J. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2019, 2, 143. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Baek, D.; Cha, J.Y.; Liao, X.; Kang, S.H.; McClung, C.R.; Lee, S.Y.; Yun, D.J.; Kim, W.Y. Hos15 interacts with the histone deacetylase hda9 and the evening complex to epigenetically regulate the floral activator gigantea. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, F.-Y.; Chen, F.-F.; Li, C.; Chen, C.; Lai, Y.-C.; Chen, J.-H.; Cui, Y.; Wu, K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018, 46, 10669–10681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Chen, J.H.; Cui, Y.; Wu, K. The LDL1/2-HDA6 histone modification complex interacts with TOC1 and regulates the core circadian clock components in Arabidopsis. Front. Plant Sci. 2019, 10, 233. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Huang, T.-Y.; Yu, H.H.; Ando, A.; Mas, P.; Ha, M.; Chen, Z.J. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 2019, 20, 170. [Google Scholar] [CrossRef]
- Ma, Y.; Gil, S.; Grasser, K.D.; Mas, P. Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in arabidopsis. Plant Cell 2018, 30, 907–924. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Hu, H.; Ren, H.; Yang, Z.; Qiu, Q.; Qi, W.; Liu, X.; Chen, X.; Cui, X.; Li, S.; et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 2019, 10, 1303. [Google Scholar] [CrossRef] [Green Version]
- Woloszynska, M.; le Gall, S.; Neyt, P.; Boccardi, T.M.; Grasser, M.; Längst, G.; Aesaert, S.; Coussens, G.; Dhondt, S.; van de Slijke, E.; et al. Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc. Natl. Acad. Sci. USA 2019, 116, 8060–8069. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Lee, K.; Ezer, D.; Cortijo, S.; Jung, J.; Charoensawan, V.; Box, M.S.; Jaeger, K.E.; Takahashi, N.; Mas, P.; et al. The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. 2020, 182, 612–625. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Raisner, R.M.; Madhani, H.D. Patterning chromatin: Form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 2006, 16, 119–124. [Google Scholar] [CrossRef] [PubMed]
- 92. Raisner, R.M.; Hartley, P.D.; Meneghini, M.D.; Bao, M.Z.; Liu, C.L.; Schreiber, S.L.; Rando, O.J.; Madhani, H.D. Histone variant H2A.Z marks the 5’ ends of both active and inactive genes in euchromatin. Cell 2005, 123, 233–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rugnone, M.L.; Soverna, A.F.; Sanchez, S.E.; Schlaen, R.G.; Hernando, C.E.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Mas, P.; et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-García, P.; Ma, Y.; Yanovsky, M.J.; Mas, P. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 5249–5253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, T.; Takeuchi, A.; Nomoto, Y.; Nakamichi, N.; Yamashino, T. The LNK1 night light-inducible and clock-regulated gene is induced also in response to warm-night through the circadian clock nighttime repressor in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e28505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishio, H.; Nagano, A.J.; Ito, T.; Suzuki, Y.; Kudoh, H. Seasonal plasticity and diel stability of H3K27me3 in natural fluctuating environments. Nat. Plants 2020, 1091–1097. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Johansson, M.; Staiger, D. Time to flower: Interplay between photoperiod and the circadian clock. J. Exp. Bot. 2015, 66, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef]
- Duren, Z.; Wang, Y.; Wang, J.; Zhao, X.-M.; Lv, L.; Li, X.; Liu, J.; Zhu, X.-G.; Chen, L.; Wang, Y. Hierarchical graphical model reveals HFR1 bridging circadian rhythm and flower development in Arabidopsis thaliana. NPJ Syst. Biol. Appl. 2019, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- You, Y.; Sawikowska, A.; Neumann, M.; Posé, D.; Capovilla, G.; Langenecker, T.; Neher, R.A.; Krajewski, P.; Schmid, M. Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nat. Commun. 2017, 8, 15120. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Z.; Liu, Y.; An, Z.; Peng, M.; Shen, W.H.; Dong, A.; Yu, Y. MRG1/2 histone methylation readers and HD2C histone deacetylase associate in repression of the florigen gene FT to set a proper flowering time in response to day-length changes. New Phytol. 2020, 227, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Wagner, D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 2015, 23, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Gao, Z.; Wang, Y.; Chen, Z.; Zhang, W.; Huang, J.; Yu, H.; He, Y. The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 2018, 95, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.A.; Morohashi, K.; Grotewold, E.; Harmer, S.L. Arabidopsis JMJD5/JMJ30 Acts Independently of LUX ARRHYTHMO Within the Plant Circadian Clock to Enable Temperature Compensation. Front. Plant Sci. 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; McCarty, D.R. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008, 11, 548–553. [Google Scholar] [CrossRef]
- Zha, P.; Liu, S.; Li, Y.; Ma, T.; Yang, L.; Jing, Y.; Lin, R. The Evening Complex and the Chromatin-Remodeling Factor PICKLE Coordinately Control Seed Dormancy by Directly Repressing DOG1 in Arabidopsis. Plant Commun. 2020, 1, 100011. [Google Scholar] [CrossRef]
- Nonogaki, H. The Long-Standing Paradox of Seed Dormancy Unfolded? Trends Plant Sci. 2019, 24, 989–998. [Google Scholar] [CrossRef]
- Sai, J.; Johnson, C.H. Different circadian oscillators control Ca(2+) fluxes and lhcb gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 11659–11663. [Google Scholar] [CrossRef] [Green Version]
- Thain, S.C.; Murtas, G.; Lynn, J.R.; McGrath, R.B.; Millar, A.J. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002, 130, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Michael, T.P.; Salome, P.A.; McClung, C.R. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci. USA 2003, 100, 6878–6883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muranaka, T.; Oyama, T. Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci. Adv. 2016, 2, e1600500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.W.; Takahashi, N.; Hirata, Y.; Ronald, J.; Porco, S.; Davis, S.J.; Nusinow, D.A.; Kay, S.A.; Mas, P. Amobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 2020, 6, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Yakir, E.; Hassidim, M.; Melamed-Book, N.; Hilman, D.; Kron, I.; Green, R.M. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011, 68, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Ukai, K.; Oyama, T. Self-arrangement of cellular circadian rhythms through phase-resetting in plant roots. Phys. Rev. E 2012, 86, 41917. [Google Scholar] [CrossRef] [PubMed]
- Thain, S.C.; Hall, A.; Millar, A.J. Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 2000, 10, 951–956. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, H.; Nakamichi, N.; Hisatsune, M.; Murase, H.; Mizuno, T. Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Phys. Rev. Lett. 2007, 99, 98102. [Google Scholar] [CrossRef]
- Wenden, B.; Toner, D.L.K.; Hodge, S.K.; Grima, R.; Millar, A.J. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc. Natl. Acad. Sci. USA 2012, 109, 6757–6762. [Google Scholar] [CrossRef] [Green Version]
- Greenwood, M.; Domijan, M.; Gould, P.D.; Hall, A.J.W.; Locke, J.C.W. Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana. PLOS Biol. 2019, 17, e3000407. [Google Scholar] [CrossRef] [Green Version]
- Gould, P.D.; Domijan, M.; Greenwood, M.; Tokuda, I.T.; Rees, H.; Kozma-Bognar, L.; Hall, A.J.; Locke, J.C. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 2018, 7, e31700. [Google Scholar] [CrossRef]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A Hierarchical Multi-oscillator Network Orchestrates the Arabidopsis Circadian System. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, A.B.; Monreal, J.A.; Nimmo, G.A.; Kelly, C.L.; Herzyk, P.; Jenkins, G.I.; Nimmo, H.G. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 2008, 322, 1832–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimmo, H.G. Entrainment of Arabidopsis roots to the light:dark cycle by light piping. Plant Cell Environ. 2018, 41, 1742–1748. [Google Scholar] [CrossRef] [Green Version]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Ren, B. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 2017, 33, 265–289. [Google Scholar] [CrossRef]
| Histone Mark | Clock/Chromatin-Related Factor | Regulated Clock Component | Reference |
|---|---|---|---|
| Acetylation | HAF2 | PRR5, LUX | [79] |
| Deacetylation | Sin3-HDAC | CCA1, PRR9 | [80] |
| ELF3-HDA9 | TOC1 | [81] | |
| EC-HDA9-HOS15 | GI | [82] | |
| HDA6-CCA1/LHY | TOC1 | [83] | |
| HDA6-TOC1 | CCA1, LHY, other clock genes | [84] | |
| Methylation | SDG2 (ATXR3) | CCA1, LHY | [78,85] |
| RVE8/LNKs | PRR5, TOC1 nascent RNAs | [86] | |
| Demethylation | JMJ14 | CCA1, LHY | [85] |
| CCA1/LHY-LDL1/2 | TOC1 | [83] | |
| TOC1-LDL1/2 | CCA1, LHY, other clock genes | [84] | |
| JMJ13 | CCA1, LHY | [87] | |
| Monoubiquitination | HUB1/HUB2 | CCA1 | [88] |
| Histone Variant H2A.Z | ELF3-SWR1 | PRR7, PRR9 | [89] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maric, A.; Mas, P. Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis. Genes 2020, 11, 1170. https://doi.org/10.3390/genes11101170
Maric A, Mas P. Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis. Genes. 2020; 11(10):1170. https://doi.org/10.3390/genes11101170
Chicago/Turabian StyleMaric, Aida, and Paloma Mas. 2020. "Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis" Genes 11, no. 10: 1170. https://doi.org/10.3390/genes11101170




