Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Sample Collection of T. radiata
2.3. Total RNA Extraction and cDNA Synthesis
2.4. Gene Cloning and Primer Design
2.5. RT-qPCR with SYBR Green
2.6. Determination of Reference Gene Expression Stability
2.7. Determination of Gene Expression Levels based on Different Reference Genes
3. Results
3.1. PCR Amplification and Performance of Candidate Reference Genes in T. radiata
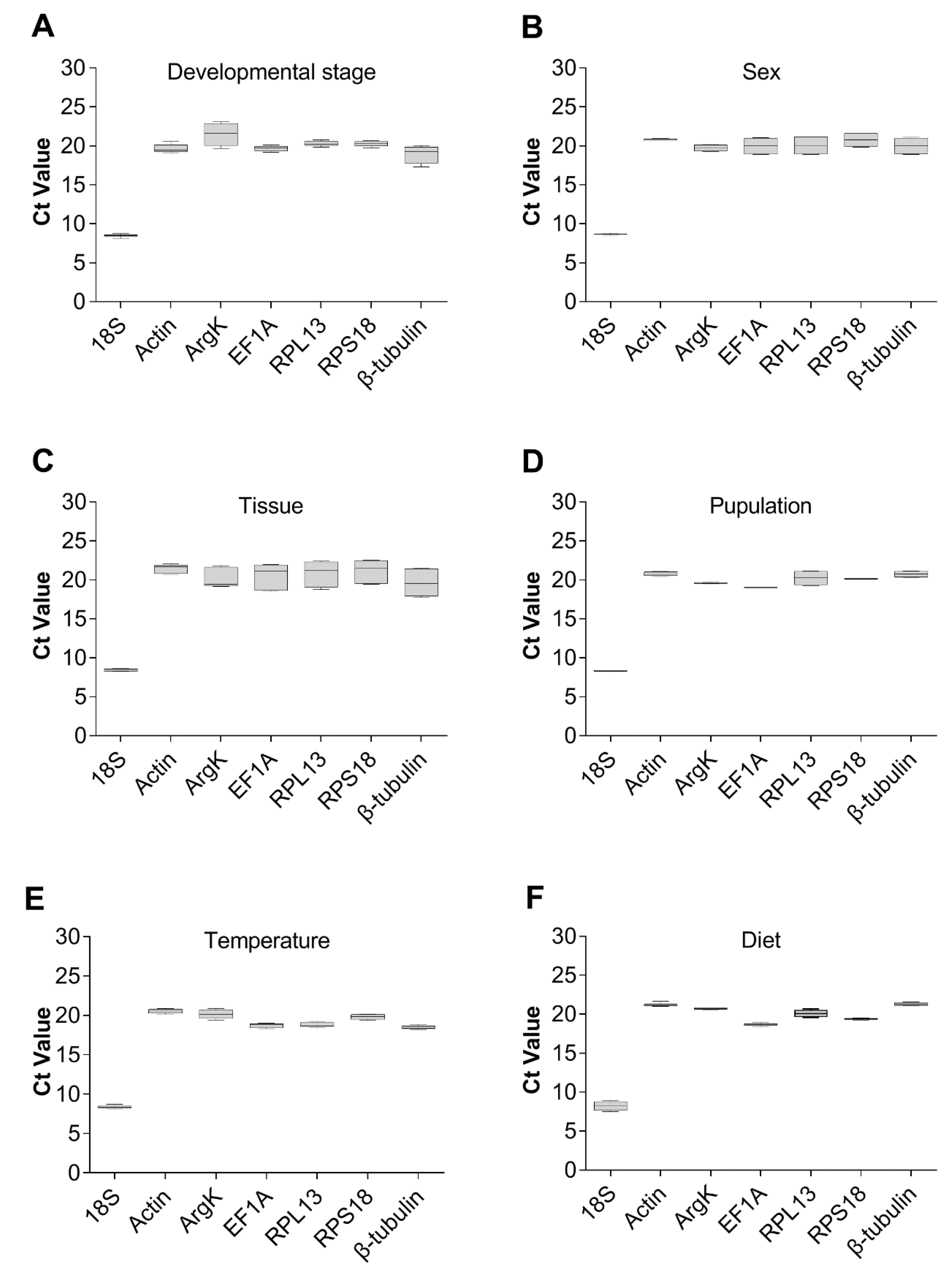
3.2. Ct Values of Reference Genes
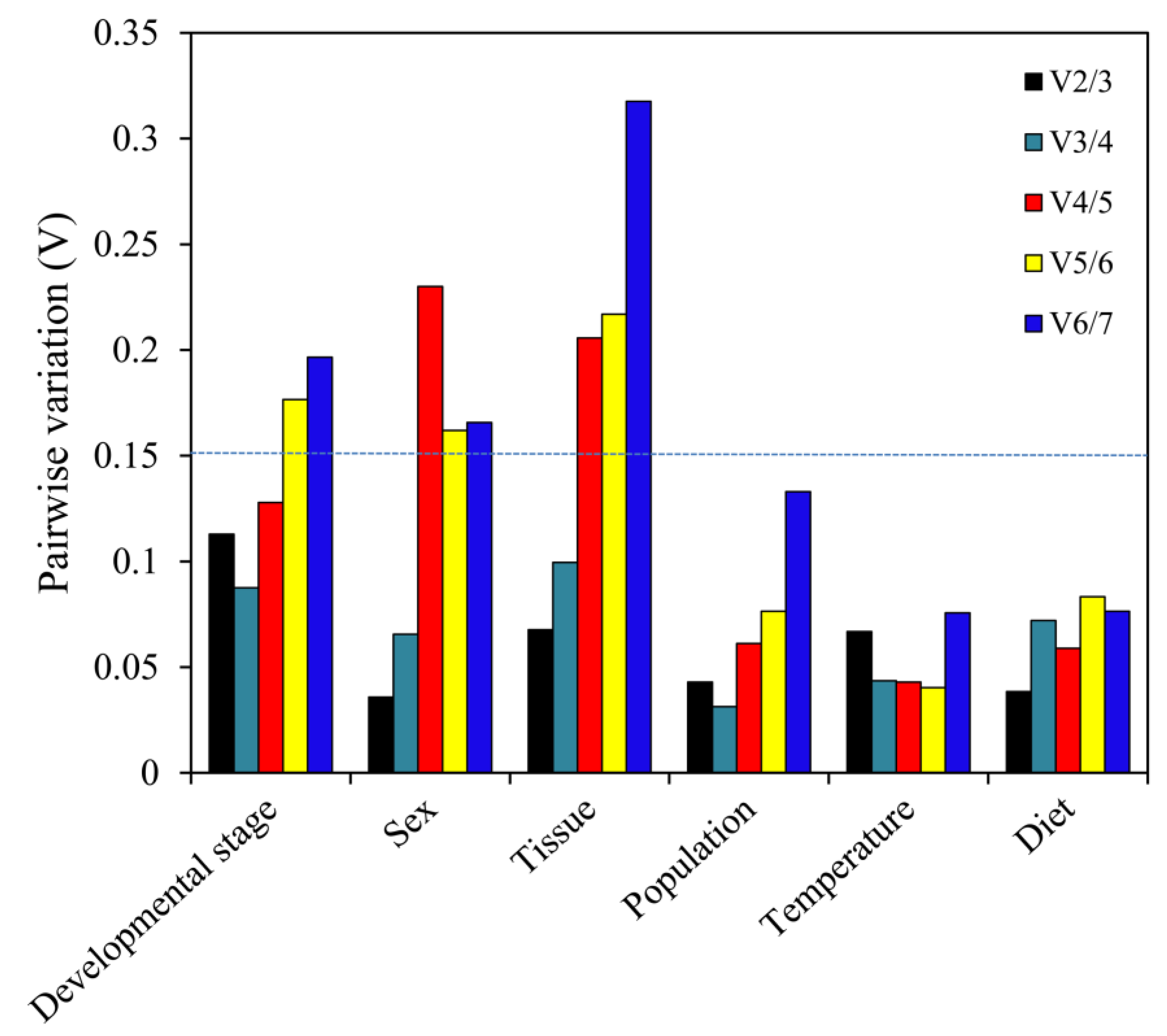
3.3. Stability of the Reference Genes under Specific Experimental Conditions
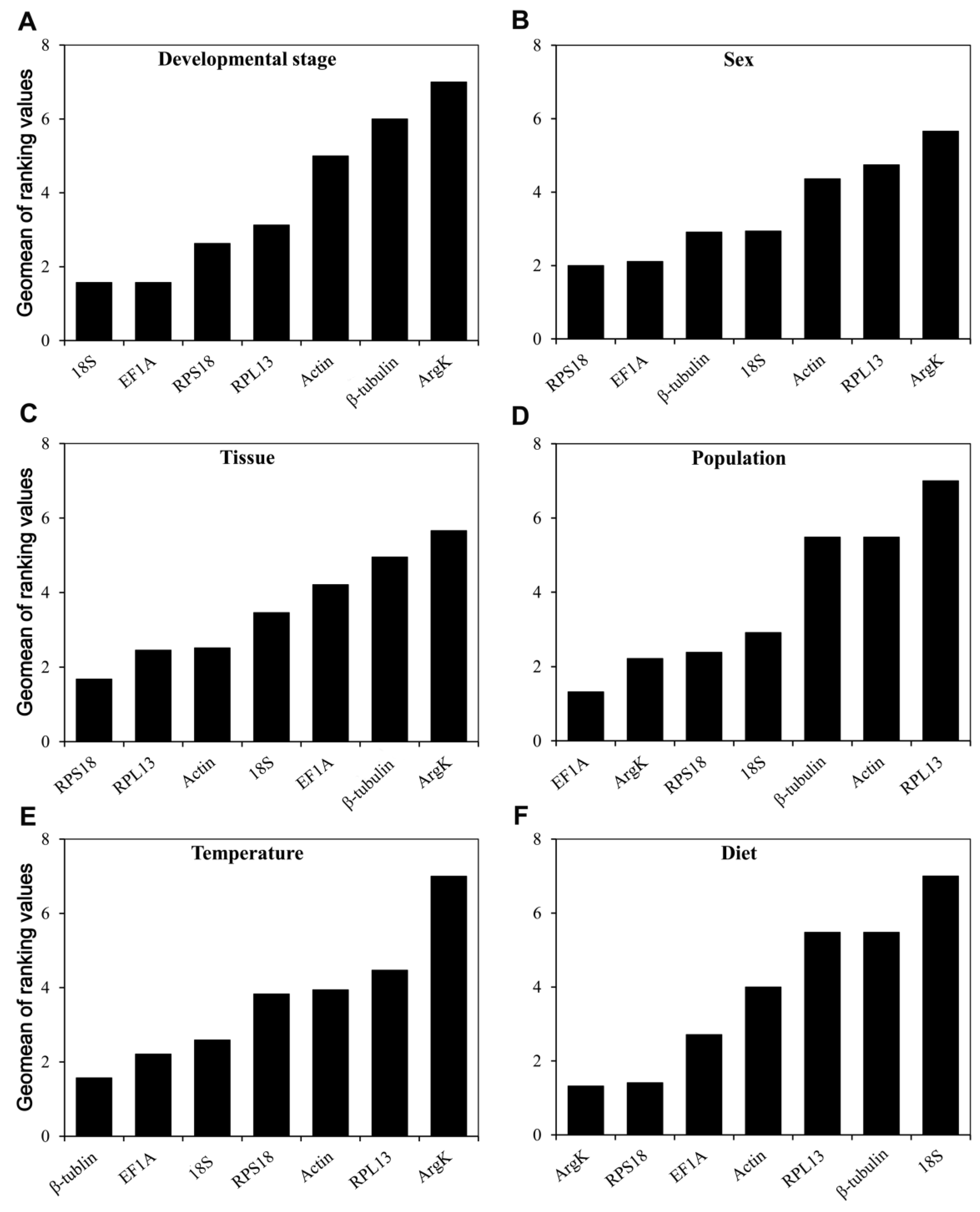
3.4. The Overall RefFinder Ranking for Reference Gene Expression Stability
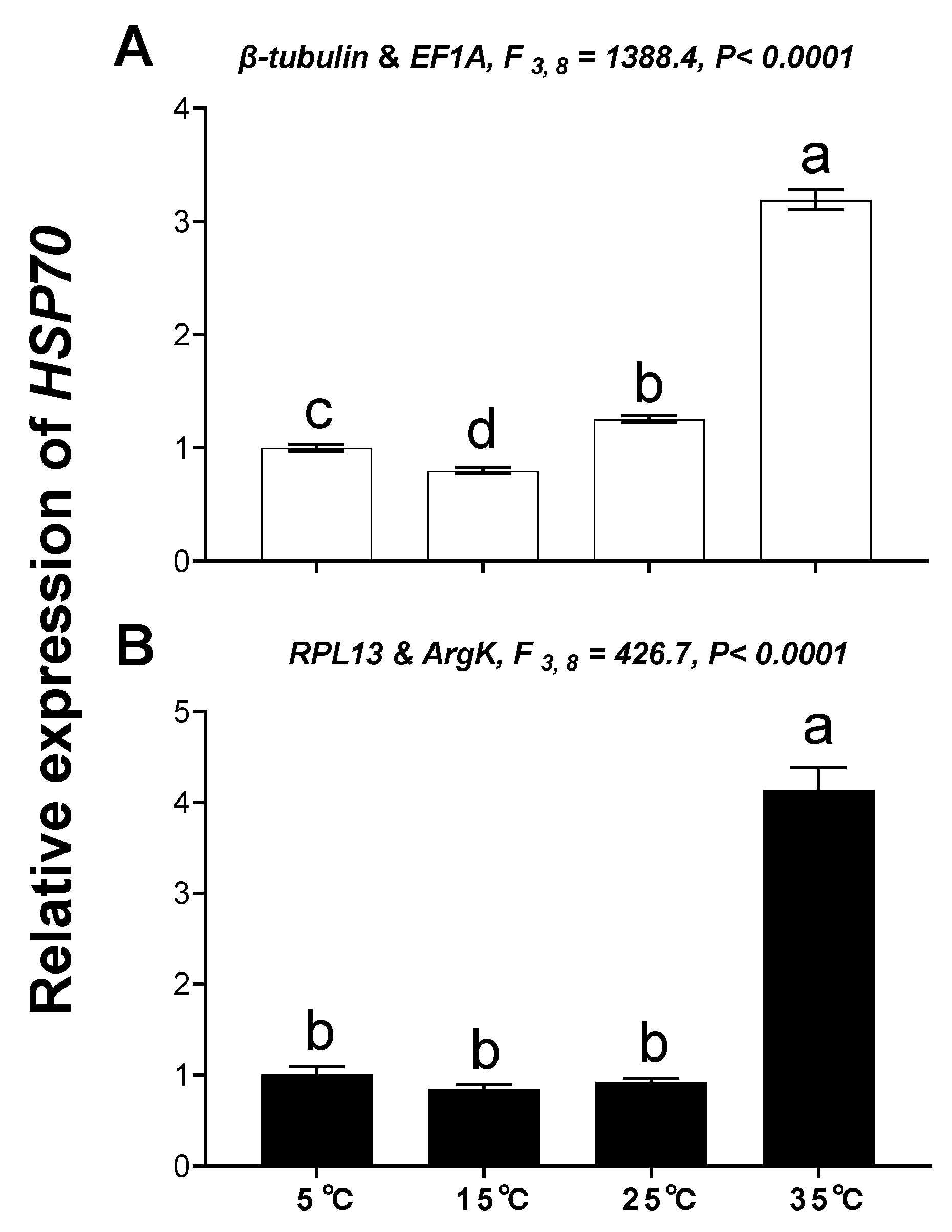
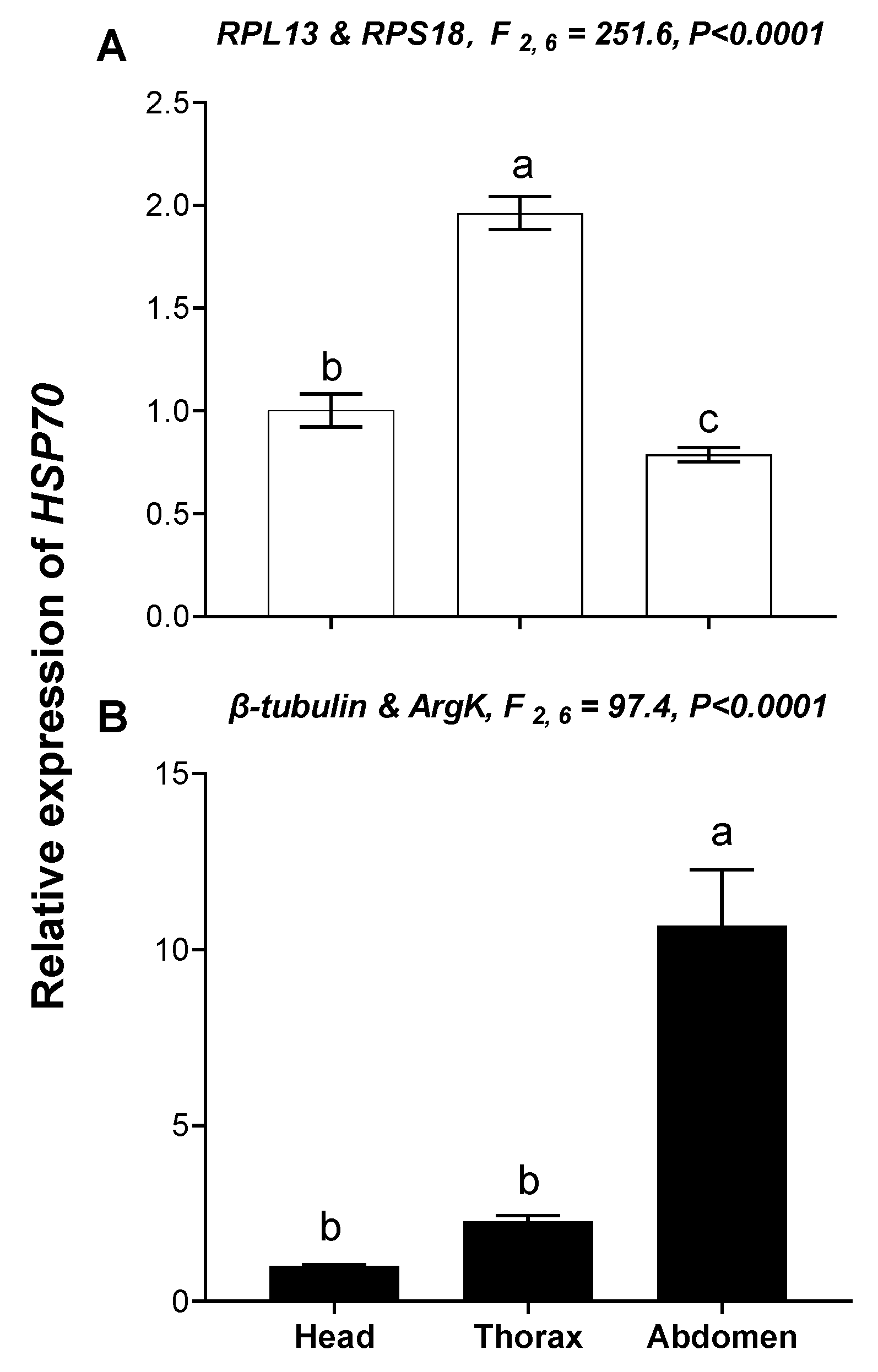
3.5. Validation of the Selected Reference Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bove, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.Z.; Li, J.Y.; Rangel, L.T.; Martins, J.J. The Candidatus Liberibacter-host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y. The status of citrus Huanglongbing in China. Trop. Plant Pathol. 2020, 45, 279–284. [Google Scholar] [CrossRef]
- Bassanezi, R.B.; Lopes, S.A.; De Miranda, M.P.; Wulff, N.A.; Volpe, H.X.L.; Ayres, A.J. Overview of citrus huanglongbing spread and management strategies in Brazil. Trop. Plant Pathol. 2020, 45, 251–264. [Google Scholar] [CrossRef]
- Grafton-Cardwell, E.E.; Stelinski, L.L.; Stansly, P.A. Biology and management of asian citrus psyllid, vector of the Huanglongbing pathogens. Annu. Rev. Entomol. 2013, 58, 413–432. [Google Scholar] [CrossRef] [Green Version]
- Halbert, S.E.; Manjunath, K.L. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: A literature review and assessment of risk in Florida. Fla. Entomol. 2004, 87, 330–353. [Google Scholar] [CrossRef]
- Yang, Y.P.; Huang, M.D.; Beattie, G.a.C.; Xia, Y.L.; Ouyang, G.C.; Xiong, J.J. Distribution, biology, ecology and control of the psyllid Diaphorina citri Kuwayama, a major pest of citrus: A status report for China. Int. J. Pest Manag. 2006, 52, 343–352. [Google Scholar] [CrossRef]
- Tiwari, S.; Mann, R.S.; Rogers, M.E.; Stelinski, L.L. Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 2011, 67, 1258–1268. [Google Scholar] [CrossRef]
- Chen, X.D.; Stelinski, L.L. Resistance management for Asian citrus psyllid, Diaphorina citri Kuwayama, in Florida. Insects 2017, 8, 103. [Google Scholar] [CrossRef] [Green Version]
- Kanga, L.H.B.; Eason, J.; Haseeb, M.; Qureshi, J.; Stansly, P. Monitoring for insecticide resistance in Asian citrus psyllid (Hemiptera: Psyllidae) populations in Florida. J. Econ. Entomol. 2016, 109, 832–836. [Google Scholar] [CrossRef]
- Naeem, A.; Freed, S.; Jin, F.L.; Akmal, M.; Mehmood, M. Monitoring of insecticide resistance in Diaphorina citri Kuwayama (Hemiptera: Psyllidae) from citrus groves of Punjab, Pakistan. Crop Prot. 2016, 86, 62–68. [Google Scholar] [CrossRef]
- Tian, F.J.; Mo, X.F.; Rizvi, S.A.H.; Li, C.F.; Zeng, X.N. Detection and biochemical characterization of insecticide resistance in field populations of Asian citrus psyllid in Guangdong of China. Sci. Rep. 2018, 8, 12587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne, J.; Quilici, S.; Marival, D.; Franck, A. Biological control of Diaphorina citri (Hemiptera: Psyllidae) in Guadeloupe by imported Tamarixia radiata (Hymenoptera: Eulophidae). Fruits 2001, 56, 307–315. [Google Scholar] [CrossRef] [Green Version]
- Parra, J.R.P.; Alves, G.R.; Diniz, A.J.F.; Vieira, J.M. Tamarixia radiata (Hymenoptera: Eulophidae) x Diaphorina citri (Hemiptera: Liviidae): Mass rearing and potential use of the parasitoid in Brazil. J. Integr. Pest Manag. 2016, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Chavez, Y.; Chirinos, D.T.; Gonzalez, G.; Lemos, N.; Fuentes, A.; Castro, R.; Kondo, T. Tamarixia radiata (Waterston) and Cheilomenes sexmaculata (Fabricius) as biological control agents of Diaphorina citri Kuwayama in Ecuador. Chil. J. Agric. Res. 2017, 77, 180–184. [Google Scholar] [CrossRef] [Green Version]
- Diniz, A.J.F.; Garcia, A.G.; Alves, G.R.; Reigada, C.; Vieira, J.M.; Parra, J.R.P. The enemy is outside: Releasing the parasitoid Tamarixia radiata (Hymenoptera: Eulophidae) in external sources of HLB inocula to control the Asian citrus psyllid Diaphorina citri (Hemiptera: Liviidae). Neotrop. Entomol. 2020, 49, 250–257. [Google Scholar] [CrossRef]
- Flores, D.; Ciomperlik, M. Biological control using the ectoparasitoid, Tamarixia radiata, against the Asian citrus psyllid, Diaphorina citri, in the Lower Rio Grande Valley of Texas. Southw. Entomol. 2017, 42, 49–59. [Google Scholar] [CrossRef]
- Patt, J.M.; Rohrig, E. Laboratory evaluations of the foraging success of Tamarixia radiata (Hymenoptera: Eulophidae) on flowers and extrafloral nectaries: Potential use of nectar plants for conservation biological control of Asian citrus psyllid (Hemiptera: Liviidae). Fla. Entomol. 2017, 100, 149–156. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.H.; Lu, Z.T.; Guo, C.F.; Shen, Z.L.; Wang, Z.Q.; Sang, W.; Qiu, B.L. Effects of cold storage on the fitness of Tamarixia radiata, a dominant parasitoid of Asian citrus psyllid Diaphorina citri. Crop Prot. 2020, 128, 104988. [Google Scholar] [CrossRef]
- Teng, Z.W.; Xiong, S.J.; Xu, G.; Gan, S.Y.; Chen, X.; Stanley, D.; Yan, Z.C.; Ye, G.Y.; Fang, Q. Protein discovery: Combined transcriptomic and proteomic analyses of venom from the endoparasitoid Cotesia chilonis (Hymenoptera: Braconidae). Toxins 2017, 9, 135. [Google Scholar] [CrossRef] [Green Version]
- Fang, Q.; Wang, B.B.; Ye, X.H.; Wang, F.; Ye, G.Y. Venom of parasitoid pteromalus puparum impairs host humoral antimicrobial activity by decreasing host cecropin and lysozyme gene expression. Toxins 2016, 8, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Lin, Z.; Fang, Q.; Wang, J.L.; Yan, Z.C.; Zou, Z.; Song, Q.S.; Ye, G.Y. The genomic and transcriptomic analyses of serine proteases and their homologs in an endoparasitoid, Pteromalus puparum. Dev. Comp. Immunol. 2017, 77, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mei, Y.T.; Fang, Q.; Wang, J.L.; Yan, Z.C.; Song, Q.S.; Lin, Z.; Ye, G.Y. Identification and characterization of serine protease inhibitors in a parasitic wasp, Pteromalus puparum. Sci. Rep. 2017, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Stansly, P.A. Biology of Tamarixia radiata (Hymenoptera: Eulophidae), parasitoid of the citrus greening disease vector Diaphorina citri (Hemiptera: Psylloidea): A mini review. Fla. Entomol. 2014, 97, 1404–1413. [Google Scholar] [CrossRef]
- Du, Y.M.; Song, X.; Liu, X.J.; Ouyang, Z.G.; Lu, Z.J. Mitochondrial genome of Tamarixia radiata (Hymenoptera: Chalcidoidea: Eulophidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 2019, 4, 2839–2840. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.K.; Zhang, S.; Luo, J.Y.; Wang, C.Y.; Lu, L.M.; Zhang, L.J.; Zhu, X.Z.; Wang, L.; Lu, H.; Cui, J.J. Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene 2017, 637, 211–218. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Provenano, M.; Mocellin, S. Complementary techniques: Validation of gene expression data by quantitative real time PCR. Microarray Technol. Cancer Gene Prof. 2007, 593, 66–73. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.X.; Pan, H.P.; Liu, Y.; Zhou, X.G. Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS ONE 2014, 9, e110454. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.X.; Pan, H.P.; Liu, Y.; Zhou, X.G. Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS ONE 2015, 10, e110454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.X.; Pan, H.P.; Noland, J.E.; Zhang, D.Y.; Zhang, Z.H.; Liu, Y.; Zhou, X.G. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 2015, 5, 18201. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Chen, S.M.; Guo, M.J.; Ye, C.Y.; Qiu, B.L.; Yang, C.X.; Pan, H.P. Selection of appropriate reference genes for RT-qPCR analysis in Propylea japonica (Coleoptera: Coccinellidae). PLoS ONE 2018, 13, e0208027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.W.; Pan, H.P.; Yuan, L.; Zhou, X.G. Reference gene selection for RTqPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018, 8, 2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin, S.Y.; Pu, X.H.; Shu, B.S.; Kang, C.; Luo, S.M.; Tang, Y.; Wu, Z.Z.; Lin, J.T. Selection of reference genes for optimal normalization of quantitative real-time polymerase chain reaction results for Diaphorina citri Adults. J. Econ. Entomol. 2019, 112, 355–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, J.; Yang, C.X.; Zhang, Y.; Pan, H.P. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef] [Green Version]
- Li, R.M.; Xie, W.; Wang, S.L.; Wu, Q.J.; Yang, N.N.; Yang, X.; Pan, H.P.; Zhou, X.M.; Bai, L.Y.; Xu, B.Y.; et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 2013, 8, e53006. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhou, Y.T.; Guo, C.F.; Ou, D.; Qureshi, J.A.; Sang, W.; Qiu, B.L. Temperature and host age effects on the life history of Tamarixia radiata, a dominant parasitoid of citrus psyllid Diaphorina citri. Crop Prot. 2018, 114, 32–38. [Google Scholar] [CrossRef]
- Liu, Y.M.; Guo, S.H.; Wang, F.F.; Zhang, L.H.; Guo, C.F.; Cuthbertson, A.G.S.; Qiu, B.L.; Sang, W. Tamarixia radiata behaviour is influenced by volatiles from both plants and Diaphorina citri nymphs. Insects 2019, 10, 141. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 0034.1. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morammazi, S.; Shokrollahi, B. The pattern of HSP70 gene expression, flight activity and temperature in Apis mellifera meda Colonies. J. Therm. Biol. 2020, 19, 102647. [Google Scholar] [CrossRef]
- Pan, L.N.; Wang, F.Z.; Zhang, X.Y.; Zhao, Y.N.; Zhu, G.P.; Li, M. Identification and characterization of heat shock proteins in a parasitic wasp Chouioia cuneae (Hymenoptera: Eulophidae). Entomol. Res. 2017, 48, 145–155. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2—ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hellemans, J.; Vandesompele, J. Selection of reliable reference genes for RT-qPCR Analysis. Quantitative Real-Time PCR; Biassoni, R., Rasos, A., Eds.; Methods Protoc.: New York, NY, USA, 2014; pp. 19–26. [Google Scholar]
- Collins, C.; Patel, M.V.; Colvin, J.; Bailey, D.; Seal, S. Identification and evaluation of suitable reference genes for gene expression studies in the whitefly Bemisia tabaci (Asia I) by reverse transcription quantitative real-time PCR. J. Insect Sci. 2014, 14, 1–25. [Google Scholar] [CrossRef]
- Dai, T.M.; Lu, Z.C.; Liu, W.X.; Wan, F.H. Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PLoS ONE 2017, 12, e0173821. [Google Scholar] [CrossRef]
- Liang, P.; Guo, Y.J.; Zhou, X.G.; Gao, X.W. Expression profiling in Bemisia tabaci under insecticide treatment: Indicating the necessity for custom reference gene selection. PLoS ONE 2014, 9, e87514. [Google Scholar] [CrossRef]
- Lv, Z.H.; Pan, H.P.; Zhang, W.; Ding, T.B.; Chu, D. Reference gene selection for RT-qPCR analysis in two invasive whiteflies after the acquisition of vectored or non-vectored viruses. J. Asia Pac. Entomol. 2018, 21, 19–24. [Google Scholar] [CrossRef]
- Su, Y.L.; He, W.B.; Wang, J.; Li, J.M.; Liu, S.S.; Wang, X.W. Selection of endogenous reference genes for gene expression analysis in the Mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 2013, 106, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, J.; Chen, S.M.; Guo, M.J.; Ye, C.Y.; Qiu, B.L.; Wu, J.H.; Yang, C.X.; Pan, H.P. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 2018, 9, 1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.X.; Preisser, E.L.; Zhang, H.J.; Liu, Y.; Dai, L.Y.; Pan, H.P.; Zhou, X.G. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 2016, 7, 1672. [Google Scholar]
- Zhai, Y.F.; Lin, Q.C.; Zhou, X.H.; Zhang, X.Y.; Liu, T.L.; Yu, Y. Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae). PLoS ONE 2014, 9, e106800. [Google Scholar] [CrossRef]
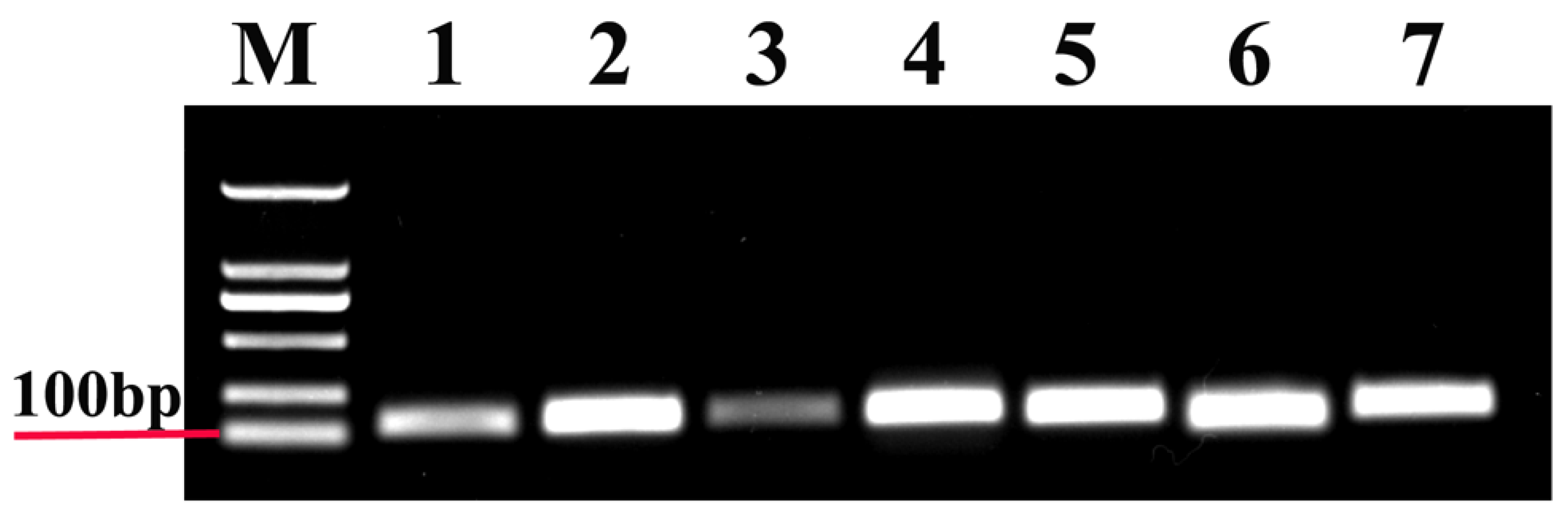
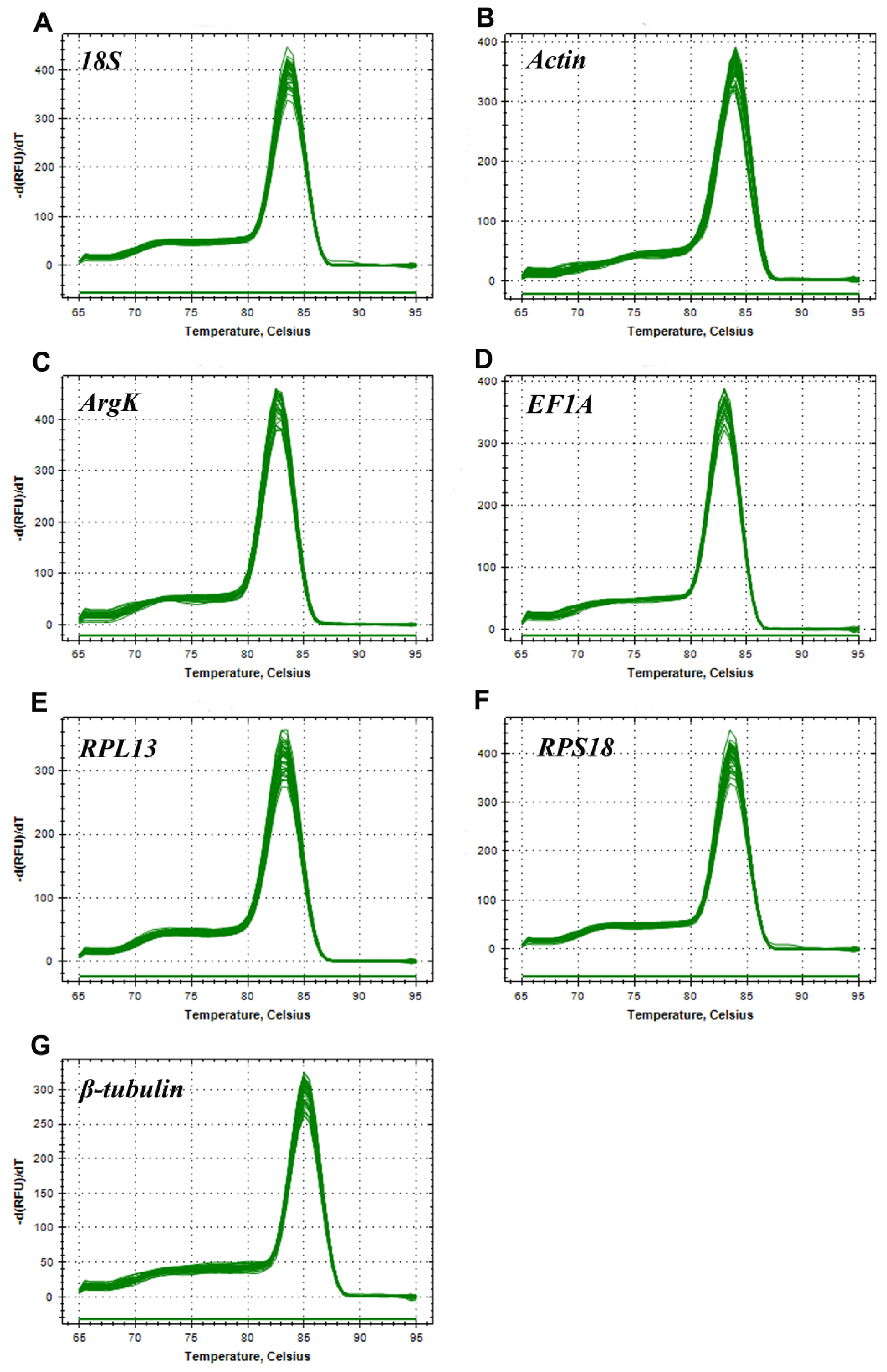
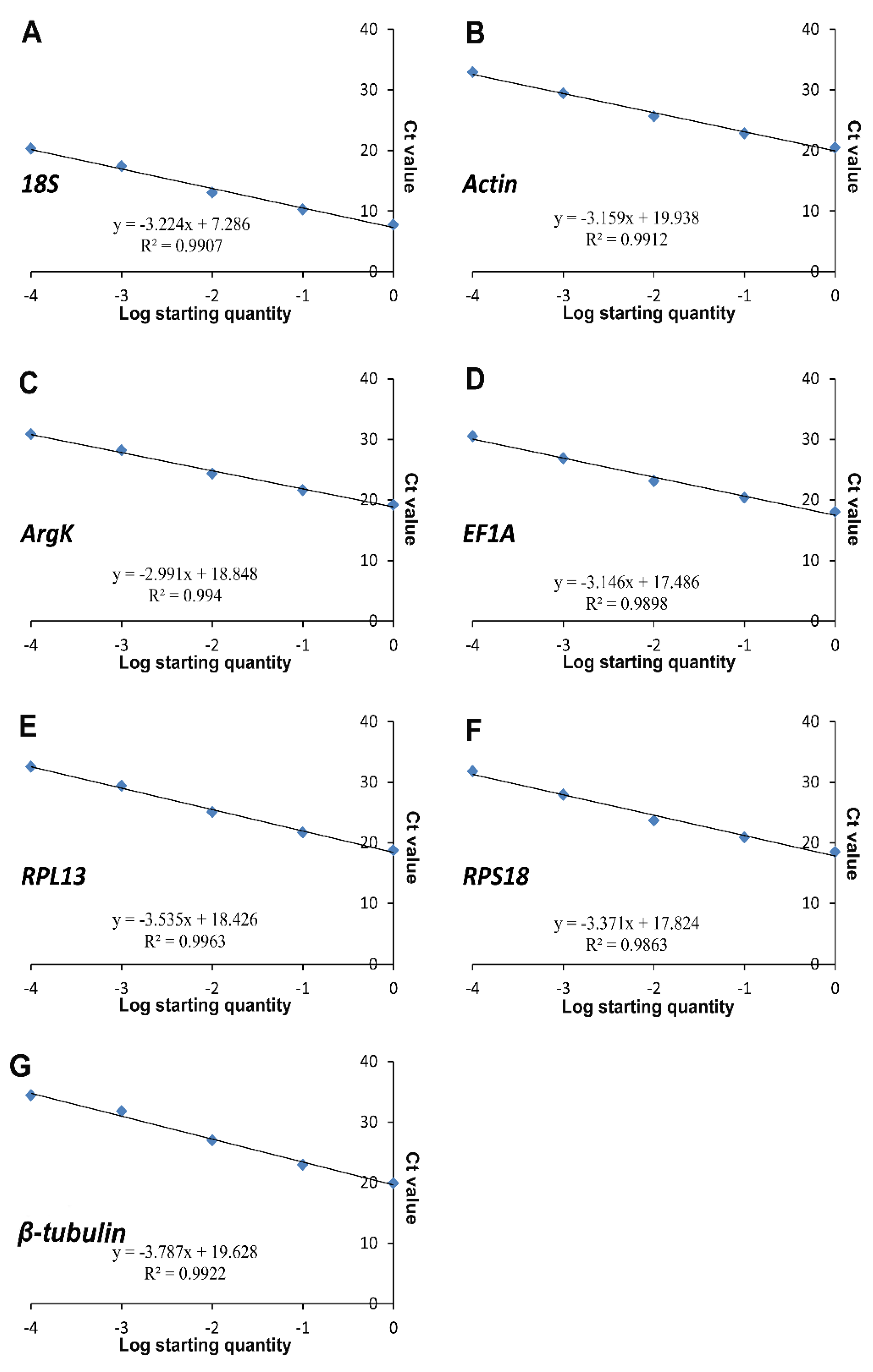
| Gene | Primer Sequences (5′–3′) | Length (bp) | Efficiency (%) | R2 | Linear Regression |
|---|---|---|---|---|---|
| 18S-F | CTTTCAAATGTCTGCCTTAT | 134 | 104.26 | 0.9907 | y = −3.224x + 7.286 |
| 18S-R | GCTGCCTTCCTTGGATGTGGT | ||||
| Actin-F | GTGTGGTGCCATATCTTCTCC | 137 | 107.28 | 0.9912 | y = −3.159x + 19.938 |
| Actin-R | ATGGTGGGTATGGGACAGAA | ||||
| ArgK-F | AGGTCTTGTCCTCGTTGTGG | 148 | 115.94 | 0.9940 | y = −2.991x + 18.848 |
| ArgK-R | CCATTAACCGGAATGAGCAA | ||||
| EF1A -F | TTCCTTCAAGTATGCCTGGG | 139 | 107.90 | 0.9898 | y = −3.146x + 17.486 |
| EF1A -R | GAAATCTCTGTGTCCAGGGG | ||||
| RPL13-F | TAGCCTTTTTGACCGGCAC | 90 | 91.82 | 0.9963 | y = −3.535x + 18.426 |
| RPL13-R | GCACAAGGGTCTCAGAAAGG | ||||
| RPS18-F | ACAAAAGGACATTGTTGACGG | 137 | 97.99 | 0.9863 | y = −3.371x + 17.824 |
| RPS18-R | GAACACGGAGACCCCAGTAG | ||||
| β-tubulin-F | GAGCAGATGCTCAACATCCA | 134 | 83.68 | 0.9922 | y = −3.787x + 19.628 |
| β-tubulin-R | GTGCTGTTTCCGATGAAGGT |
| Conditions | Rank | geNorm | NormFinder | BestKeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | ||
| Developmental stage | 1 | EF1A | 0.306 | EF1A | 0.054 | 18S | 0.12 | 18S | 0.66 |
| 2 | RPS18 | 0.306 | 18S | 0.182 | RPL13 | 0.24 | EF1A | 0.68 | |
| 3 | 18S | 0.35 | RPL13 | 0.268 | EF1A | 0.26 | RPS18 | 0.73 | |
| 4 | RPL13 | 0.374 | RPS18 | 0.337 | RPS18 | 0.27 | RPL13 | 0.73 | |
| 5 | Actin | 0.492 | Actin | 0.699 | Actin | 0.39 | Actin | 0.91 | |
| 6 | β-tubulin | 0.696 | β-tubulin | 1.066 | β-tubulin | 0.88 | β-tubulin | 1.21 | |
| 7 | ArgK | 0.907 | ArgK | 1.358 | ArgK | 1.23 | ArgK | 1.44 | |
| Sex | 1 | EF1A | 0.07 | RPS18 | 0.311 | 18S | 0.06 | RPS18 | 0.69 |
| 2 | β-tubuulin | 0.07 | EF1A | 0.607 | Actin | 0.09 | EF1A | 0.73 | |
| 3 | RPL13 | 0.098 | 18S | 0.626 | ArgK | 0.41 | β-tubulin | 0.74 | |
| 4 | RPS18 | 0.182 | β-tubulin | 0.629 | RPS18 | 0.83 | RPL13 | 0.8 | |
| 5 | 18S | 0.57 | Actin | 0.692 | EF1A | 1.02 | 18S | 0.85 | |
| 6 | Actin | 0.707 | RPL13 | 0.745 | β-tubulin | 1.04 | Actin | 0.88 | |
| 7 | ArgK | 0.837 | ArgK | 1.132 | RPL13 | 1.12 | ArgK | 1.16 | |
| Tissue | 1 | RPL13 | 0.179 | Actin | 0.218 | 18S | 0.14 | RPS18 | 0.93 |
| 2 | RPS18 | 0.179 | RPS18 | 0.514 | Actin | 0.49 | RPL13 | 0.99 | |
| 3 | EF1A | 0.207 | RPL13 | 0.753 | ArgK | 1.08 | EF1A | 1.04 | |
| 4 | β-tubulin | 0.308 | 18S | 0.779 | RPS18 | 1.12 | Actin | 1.1 | |
| 5 | Actin | 0.601 | EF1A | 0.821 | β-tubulin | 1.24 | β-tubulin | 1.11 | |
| 6 | 18S | 0.846 | β-tubulin | 0.863 | RPL13 | 1.25 | 18S | 1.3 | |
| 7 | ArgK | 1.244 | ArgK | 2.205 | EF1A | 1.32 | ArgK | 2.24 | |
| Population | 1 | EF1A | 0.06 | ArgK | 0.057 | EF1A | 0.01 | EF1A | 0.33 |
| 2 | RPS18 | 0.06 | 18S | 0.059 | RPS18 | 0.06 | ArgK | 0.34 | |
| 3 | 18S | 0.108 | EF1A | 0.09 | ArgK | 0.08 | 18S | 0.34 | |
| 4 | ArgK | 0.124 | RPS18 | 0.191 | 18S | 0.1 | RPS18 | 0.35 | |
| 5 | Actin | 0.199 | β-tubulin | 0.231 | Actin | 0.27 | β-tubulin | 0.48 | |
| 6 | β-tubulin | 0.286 | Actin | 0.502 | β-tubulin | 0.37 | Actin | 0.53 | |
| 7 | RPL13 | 0.471 | RPL13 | 0.926 | RPL13 | 0.87 | RPL13 | 0.93 | |
| Temperature | 1 | EF1A | 0.138 | β-tubulin | 0.12 | 18S | 0.14 | β-tubulin | 0.26 |
| 2 | RPS18 | 0.138 | Actin | 0.123 | β-tubulin | 0.18 | EF1A | 0.28 | |
| 3 | β-tubulin | 0.187 | 18S | 0.133 | EF1A | 0.22 | 18S | 0.28 | |
| 4 | RPL13 | 0.193 | EF1A | 0.163 | Actin | 0.24 | RPL13 | 0.28 | |
| 5 | 18S | 0.212 | RPL13 | 0.188 | RPL13 | 0.24 | Actin | 0.29 | |
| 6 | Actin | 0.233 | RPS18 | 0.254 | RPS18 | 0.29 | RPS18 | 0.31 | |
| 7 | ArgK | 0.322 | ArgK | 0.526 | ArgK | 0.42 | ArgK | 0.54 | |
| Diet | 1 | ArgK | 0.063 | ArgK | 0.032 | ArgK | 0.08 | RPS18 | 0.28 |
| 2 | RPS18 | 0.063 | RPS18 | 0.032 | RPS18 | 0.1 | EF1A | 0.3 | |
| 3 | EF1A | 0.1 | EF1A | 0.052 | EF1A | 0.12 | ArgK | 0.3 | |
| 4 | Actin | 0.197 | Actin | 0.328 | Actin | 0.16 | Actin | 0.42 | |
| 5 | β-tubulin | 0.247 | RPL13 | 0.381 | β-tubulin | 0.19 | RPL13 | 0.45 | |
| 6 | RPL13 | 0.335 | β-tubulin | 0.437 | RPL13 | 0.46 | β-tubulin | 0.48 | |
| 7 | 18S | 0.396 | 18S | 0.524 | 18S | 0.56 | 18S | 0.55 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, C.-F.; Pan, H.-P.; Zhang, L.-H.; Ou, D.; Lu, Z.-T.; Khan, M.M.; Qiu, B.-L. Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri. Genes 2020, 11, 1178. https://doi.org/10.3390/genes11101178
Guo C-F, Pan H-P, Zhang L-H, Ou D, Lu Z-T, Khan MM, Qiu B-L. Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri. Genes. 2020; 11(10):1178. https://doi.org/10.3390/genes11101178
Chicago/Turabian StyleGuo, Chang-Fei, Hui-Peng Pan, Li-He Zhang, Da Ou, Zi-Tong Lu, Muhammad Musa Khan, and Bao-Li Qiu. 2020. "Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri" Genes 11, no. 10: 1178. https://doi.org/10.3390/genes11101178






