Localization and RNA Binding of Mitochondrial Aminoacyl tRNA Synthetases
Abstract
1. Introduction
2. Mitochondrial tRNA Aminoacylation
3. Protein Import into Mitochondria
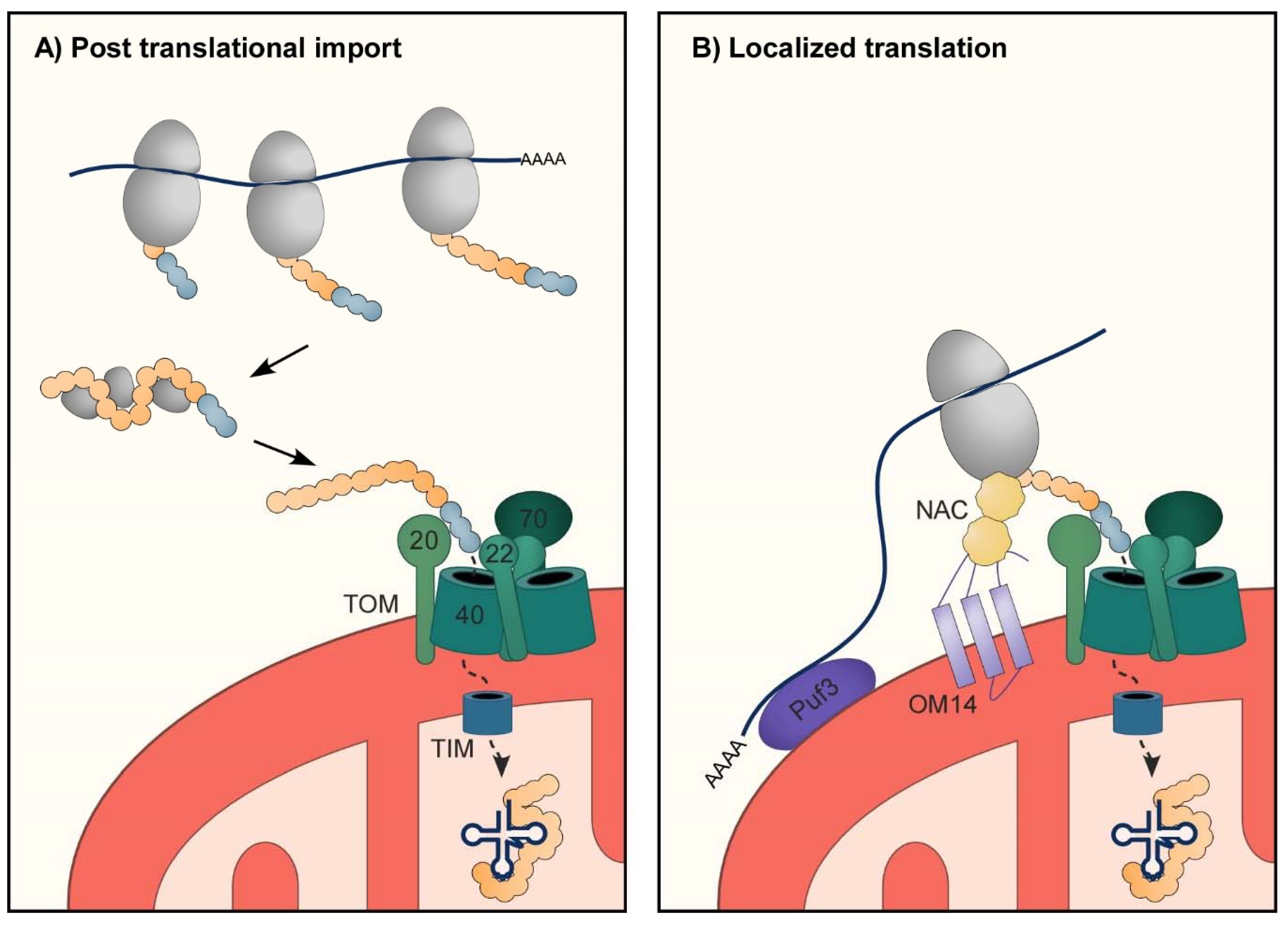
3.1. Import of mt-aaRS to Mitochondria
3.2. Mechanisms of Dual-Localization of mt-aaRSs
3.3. RNA Localization to Support Import of mt-aaRSs
4. RNA Targets of mt-aaRSs
4.1. tRNA Targeting by mt-aaRSs
4.2. Non-tRNA Targeting by mt-aaRSs
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Pizzo, P.; Filadi, R. Calcium, mitochondria and cell metabolism: A functional triangle in bioenergetics. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Vögtle, F.N.; Burkhart, J.M.; Gonczarowska-Jorge, H.; Kücükköse, C.; Taskin, A.A.; Kopczynski, D.; Ahrends, R.; Mossmann, D.; Sickmann, A.; Zahedi, R.P.; et al. Landscape of submitochondrial protein distribution. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef]
- Zardoya, R. Recent advances in understanding mitochondrial genome diversity. F1000Research 2020, 9, 270. [Google Scholar] [CrossRef]
- Daley, D.O.; Clifton, R.; Whelan, J. Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2002, 99, 10510–10515. [Google Scholar] [CrossRef]
- Von Heijne, G. Why mitochondria need a genome. FEBS Lett. 1986, 198, 1–4. [Google Scholar] [CrossRef]
- Allen, J.F. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 1993, 165, 609–631. [Google Scholar] [CrossRef]
- Cusimano, N.; Wicke, S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016, 210, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Race, H.L.; Herrmann, R.G.; Martin, W. Why have organelles retained genomes? Trends Genet. 1999, 15, 364–370. [Google Scholar] [CrossRef]
- Jacobs, H.T. Structural similarities between a mitochondrially encoded polypeptide and a family of prokaryotic respiratory toxins involved in plasmid maintenance suggest a novel mechanism for the evolutionary maintenance of mitochondrial DNA. J. Mol. Evol. 1991, 32, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Björkholm, P.; Harish, A.; Hagström, E.; Ernst, A.M.; Andersson, S.G.E. Mitochondrial genomes are retained by selective constraints on protein targeting. Proc. Natl. Acad. Sci. USA 2015, 112, 10154–10161. [Google Scholar] [CrossRef]
- Bieri, P.; Greber, B.J.; Ban, N. High-resolution structures of mitochondrial ribosomes and their functional implications. Curr. Opin. Struct. Biol. 2018, 49, 44–53. [Google Scholar] [CrossRef]
- D’Souza, A.R.; Minczuk, M. Mitochondrial transcription and translation: Overview. Essays Biochem. 2018, 62, 309–320. [Google Scholar]
- Waltz, F.; Giegé, P. Striking diversity of mitochondria-specific translation processes across eukaryotes. Trends Biochem. Sci. 2020, 45, 149–162. [Google Scholar] [CrossRef]
- Chaliotis, A.; Vlastaridis, P.; Mossialos, D.; Ibba, M.; Becker, H.D.; Stathopoulos, C.; Amoutzias, G.D. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2017, 45, 1059–1068. [Google Scholar] [CrossRef]
- Rajendran, V.; Kalita, P.; Shukla, H.; Kumar, A.; Tripathi, T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. [Google Scholar] [CrossRef]
- Suzuki, T.T.; Nagao, A.; Suzuki, T.T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef]
- González-Serrano, L.E.; Chihade, J.W.; Sissler, M. When a common biological role does not imply common disease outcomes: Disparate pathology linked to human mitochondrial aminoacyl-tRNA synthetases. J. Biol. Chem. 2019, 294, 5309–5320. [Google Scholar] [CrossRef] [PubMed]
- Duchêne, A.M.; Giritch, A.; Hoffmann, B.; Cognat, V.; Lancelin, D.; Peeters, N.M.; Zaepfel, M.; Maréchal-Drouard, L.; Small, I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 16484–16489. [Google Scholar] [CrossRef] [PubMed]
- Adrion, J.R.; White, P.S.; Montooth, K.L. The roles of compensatory evolution and constraint in aminoacyl trna synthetase evolution. Mol. Biol. Evol. 2016, 33, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Brindefalk, B.; Viklund, J.; Larsson, D.; Thollesson, M.; Andersson, S.G.E. Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol. Biol. Evol. 2007, 24, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Peterson, K.M.; Simonović, I.; Cho, C.; Söll, D.; Simonović, M. Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment. Proc. Natl. Acad. Sci. USA 2012, 109, 3281–3286. [Google Scholar] [CrossRef]
- Chimnaronk, S.; Jeppesen, M.G.; Suzuki, T.; Nyborg, J.; Watanabe, K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005, 24, 3369–3379. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Song, J.; Pfanner, N. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef]
- Grevel, A.; Pfanner, N.; Becker, T. Coupling of import and assembly pathways in mitochondrial protein biogenesis. Biol. Chem. 2019, 401. [Google Scholar] [CrossRef]
- Harbauer, A.B.; Zahedi, R.P.; Sickmann, A.; Pfanner, N.; Meisinger, C. The protein import machinery of mitochondria—A regulatory hub in metabolism, stress, and disease. Cell Metab. 2014, 19, 357–372. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Golani-Armon, A.; Arava, Y. Localization of nuclear-encoded mRNAs to mitochondria outer surface. Biochimie 2016, 81, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Saint-Georges, Y.; Garcia, M.; Delaveau, T.; Jourdren, L.; Le Crom, S.; Lemoine, S.; Tanty, V.; Devaux, F.; Jacq, C. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 2008, 3, e2293. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.C.; Jan, C.H.; Weissman, J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014, 346, 748–751. [Google Scholar] [CrossRef] [PubMed]
- Eliyahu, E.; Pnueli, L.; Melamed, D.; Scherrer, T.; Gerber, A.P.; Pines, O.; Rapaport, D.; Arava, Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol. 2010, 30, 284–294. [Google Scholar] [CrossRef]
- Vögtle, F.-N.; Wortelkamp, S.; Zahedi, R.P.; Becker, D.; Leidhold, C.; Gevaert, K.; Kellermann, J.; Voos, W.; Sickmann, A.; Pfanner, N.; et al. Global analysis of the mitochondrial n-proteome identifies a processing peptidase critical for protein stability. Cell 2009, 139, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Bausewein, T.; Mills, D.J.; Langer, J.D.; Nitschke, B.; Nussberger, S.; Kühlbrandt, W. Cryo-EM structure of the tom core complex from neurospora crassa. Cell 2017, 170, 693–700.e7. [Google Scholar] [CrossRef]
- Tucker, K.; Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019, 26, 1158–1166. [Google Scholar] [CrossRef]
- Mossmann, D.; Meisinger, C.; Vögtle, F.N. Processing of mitochondrial presequences. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 1098–1106. [Google Scholar] [CrossRef]
- Bohnert, M.; Pfanner, N.; van der Laan, M. Mitochondrial machineries for insertion of membrane proteins. Curr. Opin. Struct. Biol. 2015, 33, 92–102. [Google Scholar] [CrossRef]
- Sissler, M.; González-Serrano, L.E.; Westhof, E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 2017, 23, 693–708. [Google Scholar] [CrossRef]
- Boczonadi, V.; Jennings, M.J.; Horvath, R. The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 2018, 592, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Moulinier, L.; Ripp, R.; Castillo, G.; Poch, O.; Sissler, M. MiSynPat: An integrated knowledge base linking clinical, genetic, and structural data for disease-causing mutations in human mitochondrial aminoacyl-tRNA synthetases. Hum. Mutat. 2017, 38, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Huot, J.L.; Enkler, L.; Megel, C.; Karim, L.; Laporte, D.; Becker, H.D.; Duchêne, A.M.; Sissler, M.; Maréchal-Drouard, L. Idiosyncrasies in decoding mitochondrial genomes. Biochimie 2014, 100, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.C.; Robinson, A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016, 44, D1258–D1261. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef]
- Debard, S.; Bader, G.; De Craene, J.O.; Enkler, L.; Bär, S.; Laporte, D.; Hammann, P.; Myslinski, E.; Senger, B.; Friant, S.; et al. Nonconventional localizations of cytosolic aminoacyl-tRNA synthetases in yeast and human cells. Methods 2017, 113, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, L.; Fender, A.; Rudinger-Thirion, J.; Giegé, R.; Florentz, C.; Sissler, M. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry 2005, 44, 4805–4816. [Google Scholar] [CrossRef]
- Peikert, C.D.; Mani, J.; Morgenstern, M.; Käser, S.; Knapp, B.; Wenger, C.; Harsman, A.; Oeljeklaus, S.; Schneider, A.; Warscheid, B. Charting organellar importomes by quantitative mass spectrometry. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Carapito, C.; Kuhn, L.; Karim, L.; Rompais, M.; Rabilloud, T.; Schwenzer, H.; Sissler, M. Two proteomic methodologies for defining N-termini of mature human mitochondrial aminoacyl-tRNA synthetases. Methods 2017, 113, 111–119. [Google Scholar] [CrossRef]
- Calvo, S.E.; Julien, O.; Clauser, K.R.; Shen, H.; Kamer, K.J.; Wells, J.A.; Mootha, V.K. Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell. Proteom. 2017, 16, 512–523. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Huber, W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
- Rojas-Duran, M.F.; Gilbert, W.V. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA 2012. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Nasuno, R.; Yoshikawa, Y.; Jung, M.; Ida, T.; Matsunaga, T.; Morita, M.; Takagi, H.; Motohashi, H.; Akaike, T. Mitochondrial cysteinyl-tRNA synthetase is expressed via alternative transcriptional initiation regulated by energy metabolism in yeast cells. J. Biol. Chem. 2019, 294, 13781–13788. [Google Scholar] [CrossRef] [PubMed]
- Chatton, B.; Walter, P.; Ebel, J.P.; Lacroute, F.; Fasiolo, F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 1988, 263, 52–57. [Google Scholar] [PubMed]
- Wang, C.-C.; Chang, K.-J.; Tang, H.-L.; Hsieh, C.-J.; Schimmel, P. Mitochondrial form of a tRNA synthetase can be made bifunctional by manipulating its leader peptide †. Biochemistry 2003, 42, 1646–1651. [Google Scholar] [CrossRef]
- Natsoulis, G.; Hilger, F.; Fink, G.R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell 1986, 46, 235–243. [Google Scholar] [CrossRef]
- Chiu, M.I.; Mason, T.L.; Fink, G.R. HTS1 encodes both the cytoplasmic and mitochondrial histidyl-tRNA synthetase of Saccharomyces cerevisiae: Mutations alter the specificity of compartmentation. Genetics 1992, 132, 987–1001. [Google Scholar]
- Bader, G.; Enkler, L.; Araiso, Y.; Hemmerle, M.; Binko, K.; Baranowska, E.; De Craene, J.O.; Ruer-Laventie, J.; Pieters, J.; Tribouillard-Tanvier, D.; et al. Assigning mitochondrial localization of dual localized proteins using a yeast Bi-Genomic Mitochondrial-Split-GFP. Elife 2020, 9. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Tolkunova, E.; Park, H.; Xia, J.; King, M.P.; Davidson, E. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual: Alternative splicing of the primary transcript. J. Biol. Chem. 2000. [Google Scholar] [CrossRef] [PubMed]
- Waldron, A.L.; Cahan, S.H.; Franklyn, C.S.; Ebert, A.M. A single Danio rerio hars gene encodes both cytoplasmic and mitochondrial histidyl-tRNA synthetases. PLoS ONE 2017, 12, e0185317. [Google Scholar] [CrossRef] [PubMed]
- Rettig, J.; Wang, Y.; Schneider, A.; Ochsenreiter, T. Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 2012, 40, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.E.; Lorsch, J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012, 19, 568–576. [Google Scholar] [CrossRef]
- Dever, T.E.; Kinzy, T.G.; Pavitt, G.D. Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics 2016, 203, 65–107. [Google Scholar] [CrossRef]
- Kearse, M.G.; Wilusz, J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017. [Google Scholar] [CrossRef]
- Monteuuis, G.; Miścicka, A.; Świrski, M.; Zenad, L.; Niemitalo, O.; Wrobel, L.; Alam, J.; Chacinska, A.; Kastaniotis, A.J.; Kufel, J. Non-canonical translation initiation in yeast generates a cryptic pool of mitochondrial proteins. Nucleic Acids Res. 2019, 47, 5777–5791. [Google Scholar] [CrossRef]
- Tang, H.-L.; Yeh, L.-S.; Chen, N.-K.; Ripmaster, T.; Schimmel, P.; Wang, C.-C. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 2004, 279, 49656–49663. [Google Scholar] [CrossRef]
- Turner, R.J.; Lovato, M.; Schimmel, P. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J. Biol. Chem. 2000, 275, 27681–27688. [Google Scholar]
- Chang, K.J.; Wang, C.C. Translation initiation from a naturally occurring non-AUG codon in saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 13778–13785. [Google Scholar] [CrossRef]
- Mireau, H.; Lancelin, D.; Small, I.D. The same Arabidopsis gene encodes both cytosolic and mitochondrial alanyl-tRNA synthetases. Plant Cell 1996, 8, 1027–1039. [Google Scholar] [PubMed]
- Souciet, G.; Menand, B.; Ovesna, J.; Cosset, A.; Dietrich, A.; Wintz, H. Characterization of two bifunctional Arabdopsis thaliana genes coding for mitochondrial and cytosolic forms of valyl-tRNA synthetase and threonyl- tRNA synthetase by alternative use of two in-frame AUGs. Eur. J. Biochem. 1999, 266, 848–854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Duchêne, A.M.; Peeters, N.; Dietrich, A.; Cosset, A.; Small, I.D.; Wintz, H. Overlapping destinations for two dual targeted glycyl-tRNA synthetases in arabidopsis thaliana and phaseolus vulgaris. J. Biol. Chem. 2001. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, J.; Paulus, C.; Rudinger-Thirion, J.; Jossinet, F.; Frugier, M. Elaborate uORF/IRES features control expression and localization of human glycyl-tRNA synthetase. RNA Biol. 2015, 12, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Dinur-Mills, M.; Tal, M.; Pines, O. Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS ONE 2008. [Google Scholar] [CrossRef] [PubMed]
- Karniely, S.; Pines, O. Single translation-dual destination: Mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005, 6, 420–425. [Google Scholar] [CrossRef]
- Yogev, O.; Pines, O. Dual targeting of mitochondrial proteins: Mechanism, regulation and function. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1012–1020. [Google Scholar] [CrossRef]
- Berglund, A.K.; Spånning, E.; Biverståhl, H.; Maddalo, G.; Tellgren-Roth, C.; Mäler, L.; Glaser, E. Dual targeting to mitochondria and chloroplasts: Characterization of Thr-tRNA synthetase targeting peptide. Mol. Plant 2009, 2, 1298–1309. [Google Scholar] [CrossRef]
- Berglund, A.K.; Pujol, C.; Duchene, A.M.; Glaser, E. Defining the determinants for dual targeting of amino Acyl-tRNA synthetases to mitochondria and chloroplasts. J. Mol. Biol. 2009, 393, 803–814. [Google Scholar] [CrossRef]
- Pujol, C.; Maréchal-Drouard, L.; Duchêne, A.M. How can organellar protein n-terminal sequences be dual targeting signals? In silico analysis and mutagenesis approach. J. Mol. Biol. 2007, 369, 356–367. [Google Scholar] [CrossRef]
- Frechin, M.; Senger, B.; Braye, M.; Kern, D.; Martin, R.P.; Becker, H.D. Yeast mitochondrial Gln-tRNAGln is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009, 23, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Ludermer, S.; Schimmel, P. Construction and analysis of deletions in the amino-terminal extension of glutamine tRNA synthetase of saccharomyces cerevisiae. J. Biol. Chem. 1987, 262, 10807–10813. [Google Scholar]
- Nagao, A.; Suzuki, T.; Katoh, T.; Sakaguchi, Y.; Suzuki, T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc. Natl. Acad. Sci. USA 2009, 106, 16209–16214. [Google Scholar] [CrossRef] [PubMed]
- Frechin, M.; Enkler, L.; Tetaud, E.; Laporte, D.; Senger, B.; Blancard, C.; Hammann, P.; Bader, G.; Clauder-Münster, S.; Steinmetz, L.M.; et al. Expression of nuclear and mitochondrial genes encoding ATP synthase is synchronized by disassembly of a multisynthetase complex. Mol. Cell 2014, 56, 763–776. [Google Scholar] [CrossRef]
- Schmidt, O.; Pfanner, N.; Meisinger, C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 655–667. [Google Scholar] [CrossRef]
- Avendaño-Monsalve, M.C.; Ponce-Rojas, J.C.; Funes, S. From cytosol to mitochondria: The beginning of a protein journey. Biol. Chem. 2020, 401, 645–661. [Google Scholar] [CrossRef]
- Eliyahu, E.; Lesnik, C.; Arava, Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett. 2012, 586, 64–69. [Google Scholar] [CrossRef]
- Besse, F.; Ephrussi, A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008, 9, 971–980. [Google Scholar] [CrossRef]
- Reid, D.W.; Nicchitta, C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015, 16, 221–231. [Google Scholar] [CrossRef]
- Yogev, O.; Karniely, S.; Pines, O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 2007, 282, 29222–29229. [Google Scholar] [CrossRef]
- Lesnik, C.; Cohen, Y.; Atir-Lande, A.; Schuldiner, M.; Arava, Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 2014, 5, 5711. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Rojas, J.C.; Avendaño-Monsalve, M.C.; Yañez-Falcón, A.R.; Jaimes-Miranda, F.; Garay, E.; Torres-Quiroz, F.; DeLuna, A.; Funes, S. αβ′-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 2017, 284, 814–830. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wollmann, R.; Lindquist, S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 2002, 298, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Rane, N.S.; Kang, S.W.; Chakrabarti, O.; Feigenbaum, L.; Hegde, R.S. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev. Cell 2008, 15, 359–370. [Google Scholar] [CrossRef]
- Michaud, M.; Ubrig, E.; Filleur, S.; Erhardt, M.; Ephritikhine, G.; Marechal-Drouard, L.; Duchene, A.M. Differential targeting of VDAC3 mRNA isoforms influences mitochondria morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 8991–8996. [Google Scholar] [CrossRef] [PubMed]
- Gehrke, S.; Wu, Z.; Klinkenberg, M.; Sun, Y.; Auburger, G.; Guo, S.; Lu, B. PINK1 and parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015, 21, 95–108. [Google Scholar] [CrossRef]
- Corral-Debrinski, M.; Blugeon, C.; Jacq, C. In yeast, the 3’ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell Biol. 2000, 20, 7881–7892. [Google Scholar] [CrossRef]
- Ricart, J.; Izquierdo, J.M.; Di Liegro, C.M.; Cuezva, J.M. Assembly of the ribonucleoprotein complex containing the mRNA of the β-subunit of the mitochondrial H+-ATP synthase requires the participation of two distal cis-acting elements and a complex set of cellular trans-acting proteins. Biochem. J. 2002, 365, 417–428. [Google Scholar] [CrossRef]
- Gadir, N.; Haim-Vilmovsky, L.; Kraut-Cohen, J.; Gerst, J.E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 2011, 17, 1551–1565. [Google Scholar] [CrossRef]
- Tsuboi, T.; Viana, M.P.; Xu, F.; Yu, J.; Chanchani, R.; Arceo, X.G.; Tutucci, E.; Choi, J.; Chen, Y.S.; Singer, R.H.; et al. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. Elife 2020, 9. [Google Scholar] [CrossRef]
- Marc, P.; Margeot, A.; Devaux, F.; Blugeon, C.; Corral-Debrinski, M.; Jacq, C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002, 3, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Entelis, N.; Brandina, I.; Kamenski, P.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006, 20, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.A.T.; Hopper, A.K. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip. Rev. RNA 2011, 2, 802–817. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Giegé, T.; Giegé, R.; Giegé, P. TRNA biology in mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef] [PubMed]
- Sieber, F.; Duchêne, A.M.; Maréchal-Drouard, L. Mitochondrial RNA import. From diversity of natural mechanisms to potential applications. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2011; pp. 145–190. [Google Scholar]
- Giegé, R.; Eriani, G. Transfer RNA recognition and aminoacylation by synthetases. In ELS; John Wiley & Sons, Ltd.: Chichester, UK, 2014. [Google Scholar]
- Lynch, M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol. Biol. Evol. 1997, 14, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Kuhle, B.; Chihade, J.; Schimmel, P. Relaxed sequence constraints favor mutational freedom in idiosyncratic metazoan mitochondrial tRNAs. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Sohm, B.; Frugier, M.; Brulé, H.; Olszak, K.; Przykorska, A.; Florentz, C. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003, 328, 995–1010. [Google Scholar] [CrossRef]
- Sohm, B.; Sissler, M.; Park, H.; King, M.P.; Florentz, C. Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 2004, 339, 17–29. [Google Scholar] [CrossRef]
- Fender, A.; Sauter, C.; Messmer, M.; Pütz, J.; Giegé, R.; Florentz, C.; Sissler, M. Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 2006, 281, 15980–15986. [Google Scholar] [CrossRef]
- Bonnefond, L.; Frugier, M.; Giegé, R.; Rudinger-Thirion, J. Human mitochondrial TyrRS disobeys the tyrosine identity rules. RNA 2005, 11, 558–562. [Google Scholar] [CrossRef]
- Mcclain, W.H.; Foss, K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 1988, 240, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Florentz, C.; Kern, D.; Gangloff, J.; Eriani, G.; Moras, D. Aspartate identity of transfer RNAs. Biochimie 1996, 78, 605–623. [Google Scholar] [CrossRef]
- Helm, M.; Brulé, H.; Friede, D.; Giegé, R.; Pütz, D.; Florentz, C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 2000, 6, 1356–1379. [Google Scholar] [CrossRef] [PubMed]
- Pütz, J.; Dupuis, B.; Sissler, M.; Florentz, C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA 2007, 13, 1184–1190. [Google Scholar] [CrossRef]
- Ueda, T.; Ohta, T.; Watanabe, K. Large scale isolation and some properties of AGY-specific serine tRNA from bovine heart mitochondria. J. Biochem. 1985, 98, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Hanada, T.; Suzuki, T.; Yokogawa, T.; Takemoto-Hori, C.; Sprinzl, M.; Watanabe, K. Translation ability of mitochondrial tRNASSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells 2001, 6, 1019–1030. [Google Scholar] [CrossRef]
- Masta, S.E.; Boore, J.L. Parallel evolution of truncated transfer rna genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef]
- Wende, S.; Platzer, E.G.; Jühling, F.; Pütz, J.; Florentz, C.; Stadler, P.F.; Mörl, M. Biological evidence for the world’s smallest tRNAs. Biochimie 2014, 100, 151–158. [Google Scholar] [CrossRef]
- Jühling, F.; Pütz, J.; Florentz, C.; Stadler, P.F. Armless mitochondrial tRNAs in enoplea (nematoda). RNA Biol. 2012, 9, 1161–1166. [Google Scholar] [CrossRef]
- Chen, D.S.; Jin, P.Y.; Zhang, K.J.; Ding, X.L.; Yang, S.X.; Ju, J.F.; Zhao, J.Y.; Hong, X.Y. The complete mitochondrial genomes of six species of Tetranychus provide insights into the phylogeny and evolution of spider mites. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Li, C.; Fang, W.Y.; Yu, X.P. The complete mitochondrial genome of two tetragnatha spiders (Araneae: Tetragnathidae): Severe truncation of tRNAs and novel gene rearrangements in araneae. Int. J. Biol. Sci. 2016, 12, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Jühling, T.; Duchardt-Ferner, E.; Bonin, S.; Wöhnert, J.; Pütz, J.; Florentz, C.; Betat, H.; Sauter, C.; Mörl, M. Small but large enough: Structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax. Nucleic Acids Res. 2018, 46, 9170–9180. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Huang, X.; Deng, J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020, 158, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Bover, P.; Bidegaray-Batista, L.; Arnedo, M.A. Arm-less mitochondrial tRNAs conserved for over 30 millions of years in spiders. BMC Genom. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Neuenfeldt, A.; Lorber, B.; Ennifar, E.; Gaudry, A.; Sauter, C.; Sissler, M.; Florentz, C. Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res. 2013, 41, 2698–2708. [Google Scholar] [CrossRef]
- Klipcan, L.; Moor, N.; Finarov, I.; Kessler, N.; Sukhanova, M.; Safro, M.G. Crystal structure of human mitochondrial PheRS complexed with tRNA Phe in the active “open” state. J. Mol. Biol. 2012, 415, 527–537. [Google Scholar] [CrossRef]
- Bae Rho, S.; Martinis, S.A. The bl4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase to facilitate RNA splicing activity. RNA 2000, 6, 1882–1894. [Google Scholar]
- Sarkar, J.; Poruri, K.; Boniecki, M.T.; McTavish, K.K.; Martinis, S.A. Yeast mitochondrial leucyl-tRNA synthetase CP1 domain has functionally diverged to accommodate RNA splicing at expense of hydrolytic editing. J. Biol. Chem. 2012, 287, 14772–14781. [Google Scholar] [CrossRef]
- Labouesse, M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990, 224, 209–221. [Google Scholar] [CrossRef]
- Houman, F.; Rho, S.B.; Zhang, J.; Shen, X.; Wang, C.C.; Schimmel, P.; Martinis, S.A. A prokaryote and human tRNA synthetase provide an essential RNA splicing function in yeast mitochondria. Proc. Natl. Acad. Sci. USA 2000, 97, 13743–13748. [Google Scholar] [CrossRef] [PubMed]
- Boniecki, M.T.; Rho, S.B.; Tukalo, M.; Hsu, J.L.; Romero, E.P.; Martinis, S.A. Leucyl-tRNA synthetase-dependent and -independent activation of a Group I intron. J. Biol. Chem. 2009, 284, 26243–26250. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.A.; Lambowitz, A.M. RNA splicing in Neurospora mitochondria. Defective splicing of mitochondrial mRNA precursors in the nuclear mutant cyt18-1. J. Mol. Biol. 1985, 184, 413–428. [Google Scholar] [CrossRef]
- Kittle, J.D.; Mohr, G.; Gianelos, J.A.; Wang, H.; Lambowitz, A.M. The neurospora mitochondrial tyrosyl-tRNA synthetase is sufficient for group I intron splicing in vitro and uses the carboxy-terminal tRNA-binding domain along with other regions. Genes Dev. 1991, 5, 1009–1021. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kämper, U.; Kück, U.; Cherniack, A.D.; Lambowitz, A.M. The mitochondrial tyrosyl-tRNA synthetase of Podospora anserina is a bifunctional enzyme active in protein synthesis and RNA splicing. Mol. Cell. Biol. 1992, 12, 499–511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.; Mohr, G.; Lambowitz, A.M. The Neurosporea crassa CYT-18 protein C-terminal RNA-binding domain helps stabilize interdomain tertiary interactions in group I introns. RNA 2004, 10, 634–644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohr, G.; Rennard, R.; Cherniack, A.D.; Stryker, J.; Lambowitz, A.M. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J. Mol. Biol. 2001, 307, 75–92. [Google Scholar] [CrossRef]
- Lamech, L.T.; Mallam, A.L.; Lambowitz, A.M. Evolution of RNA-protein interactions: Non-specific binding led to rna splicing activity of fungal mitochondrial tyrosyl-tRNA synthetases. PLoS Biol. 2014, 12, e1002028. [Google Scholar] [CrossRef]
- Paukstelis, P.J.; Lambowitz, A.M. Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity. Proc. Natl. Acad. Sci. USA 2008, 105, 6010–6015. [Google Scholar] [CrossRef]
- Paukstelis, P.J.; Chen, J.H.; Chase, E.; Lambowitz, A.M.; Golden, B.L. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature 2008, 451, 94–97. [Google Scholar] [CrossRef]
- Guo, M.; Yang, X.-L.; Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010, 11, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Myers, C.A.; Kuhla, B.; Cusack, S.; Lambowitz, A.M. tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 2002, 99, 2630–2635. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; Tsang, T.H.; Buck, M.; Christman, M.F. The leader mRNA of the histidine attenuator region resembles tRNAHis: Possible general regulatory implications. Proc. Natl. Acad. Sci. USA 1983, 80, 5240–5242. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.; Jia, J.; Mukhopadhyay, R.; Willard, B.; Kinter, M.; Fox, P.L. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol. Cell 2009, 35, 164–180. [Google Scholar] [CrossRef] [PubMed]
- Frugier, M.; Giegé, R. Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. J. Mol. Biol. 2003, 331, 375–383. [Google Scholar] [CrossRef]
- Levi, O.; Arava, Y. mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol. 2019, 17. [Google Scholar] [CrossRef]
- Levi, O.; Garin, S.; Arava, Y. RNA mimicry in post-transcriptional regulation by aminoacyl tRNA synthetases. Wiley Interdiscip. Rev. RNA 2020, 11. [Google Scholar] [CrossRef]
- Romby, P.; Caillet, J.; Ebel, C.; Sacerdot, C.; Graffe, M.; Eyermann, F.; Brunel, C.; Moine, H.; Ehresmann, C.; Ehresmann, B.; et al. The expression of E.coli threonyl-tRNA synthetase is regulated at the translational level by symmetrical operator-repressor interactions. EMBO J. 1996, 15, 5976–5987. [Google Scholar] [CrossRef]
- Torres-Larios, A.; Dock-Bregeon, A.-C.; Romby, P.; Rees, B.; Sankaranarayanan, R.; Caillet, J.; Springer, M.; Ehresmann, C.; Ehresmann, B.; Moras, D. letters structural basis of translational control by escherichia coli threonyl tRNA synthetase anticodon-stem. Nat. Struct. Biol. 2002, 9, 343–347. [Google Scholar]
- Frugier, M.; Ryckelynck, M.; Giegé, R. tRNA-balanced expression of a eukaryal aminoacyl-tRNA synthetase by an mRNA-mediated pathway. EMBO Rep. 2005, 6, 860–865. [Google Scholar] [CrossRef]
- Romby, P.; Brunel, C.; Caillet, J.; Springer, M.; Grunberg-Manago, M.; Westhof, E.; Ehresmann, C.; Ehresmann, B. Molecular mimicry in translational control of E. coli threonyl-tRNA synthetase gene. Competitive inhibition in tRNA aminoacylation and operator-repressor recognition switch using tRNA identity rules. Nucleic Acids Res. 1992, 20, 5633–5640. [Google Scholar] [CrossRef] [PubMed]
- Ryckelynck, M.; Masquida, B.; Giegé, R.; Frugier, M. An intricate RNA structure with two tRNA-derived motifs directs complex formation between yeast aspartyl-tRNA synthetase and its mRNA. J. Mol. Biol. 2005, 354, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.J.; Park, S.; Nguyen, L.T.; Hwang, J.; Lee, E.-Y.; Giong, H.-K.; Lee, J.-S.; Yoon, I.; Lee, J.-H.; Kim, J.H.; et al. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat. Commun. 2019, 10, 1357. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, B.M.; Horos, R.; Fischer, B.; Castello, A.; Eichelbaum, K.; Alleaume, A.M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Matia-González, A.M.; Laing, E.E.; Gerber, A.P. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 2015, 22, 1027–1033. [Google Scholar] [CrossRef]


| Protein Location | aaRS | Yeast Gene | Localization to Mitochondria | Puf3 Dependence c | Translation Dependence d | Tom20 Dependence e | |
|---|---|---|---|---|---|---|---|
| Biochemical Fractions (MLR Value) a | Ribosome Proximity b | ||||||
| Mitochondrial | ArgRS | YHR091C (MSR1) | Yes (36) | Yes | Partial | No | NA |
| AsnRS | YCR024C (SLM5) | Yes (59.8) | Yes | Partial | No | Yes | |
| AspRS | YPL104W (MSD1) | Yes (51) | Yes | Partial | No | Yes | |
| GluRS | YOL033W (MSE1) | Yes (59) | Yes | Partial | No | No | |
| IleRS | YPL040C (ISM1) | Yes (66) | Yes | Partial | No | NA | |
| LeuRS | YLR382C (NAM2) | Yes (48.5) | Yes | Partial | No | No | |
| LysRS | YNL073W (MSK1) | Yes (36.7) | Yes | Partial | No | No | |
| MetRS | YGR171C (MSM1) | Yes (63.4) | Yes | Partial | No | NA | |
| PheRS | YPR047W (MSF1) | Yes (93.4) | Yes | Partial | No | Yes | |
| ProRS | YER087W (AIM10) | Yes (63.2) | Yes | Partial | No | NA | |
| SerRS | YHR011W (DIA4) | Yes (99.1) | Yes | Partial | No | NA | |
| ThrRS | YKL194C (MST1) | Yes (72.6) | Yes | Partial | No | NA | |
| TrpRS | YDR268W (MSW1) | Yes (87.2) | Yes | Partial | No | No | |
| TyrRS | YPL097W (MSY1) | Yes (94.2) | Yes | Partial | No | No | |
| Mitochondria & Cytosol | AlaRS | YOR335C (ALA1) | No | No | NR | NR | No |
| CysRS | YNL247W (CRS1) | NA | No | NA | NR | NA | |
| GlyRS | YBR121C (GRS1) | Low (13.1) | No | No | NR | No | |
| HisRS | YPR033C (HTS1) | No | No | NR | NR | No | |
| ValRS | YGR094W (VAS1) | Low (20.2) | No | No | NR | NA | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garin, S.; Levi, O.; Cohen, B.; Golani-Armon, A.; Arava, Y.S. Localization and RNA Binding of Mitochondrial Aminoacyl tRNA Synthetases. Genes 2020, 11, 1185. https://doi.org/10.3390/genes11101185
Garin S, Levi O, Cohen B, Golani-Armon A, Arava YS. Localization and RNA Binding of Mitochondrial Aminoacyl tRNA Synthetases. Genes. 2020; 11(10):1185. https://doi.org/10.3390/genes11101185
Chicago/Turabian StyleGarin, Shahar, Ofri Levi, Bar Cohen, Adi Golani-Armon, and Yoav S. Arava. 2020. "Localization and RNA Binding of Mitochondrial Aminoacyl tRNA Synthetases" Genes 11, no. 10: 1185. https://doi.org/10.3390/genes11101185
APA StyleGarin, S., Levi, O., Cohen, B., Golani-Armon, A., & Arava, Y. S. (2020). Localization and RNA Binding of Mitochondrial Aminoacyl tRNA Synthetases. Genes, 11(10), 1185. https://doi.org/10.3390/genes11101185







