The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits
Abstract
:1. Introduction
2. The Human Y Chromosome, a Complex Locus for Complex Trait Analysis
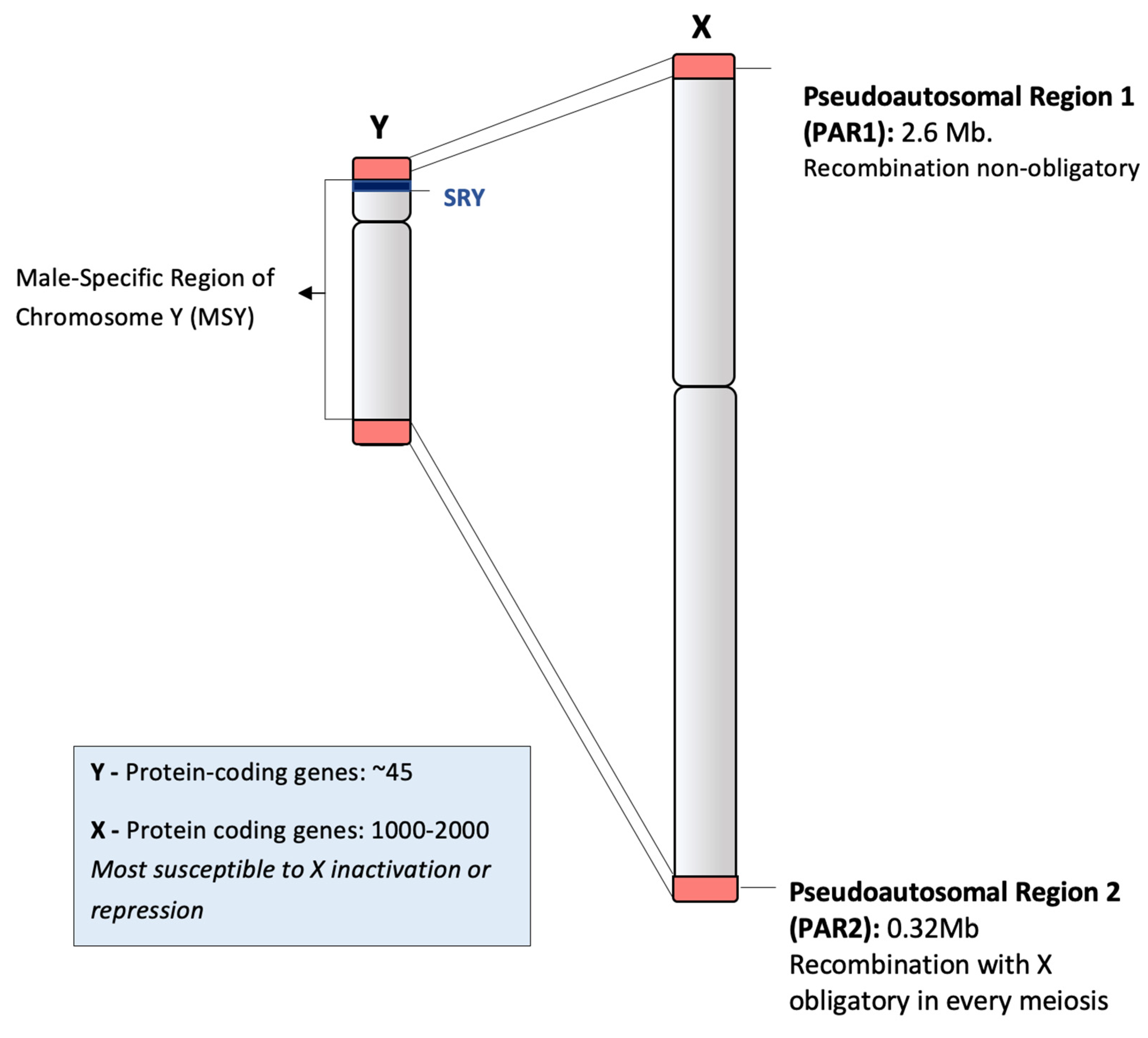
2.1. Structure of Human Chromosome Y and Recombination
2.2. The Functional Role of the Human Y Chromosome
2.2.1. Deterioration of the Y Chromosome
2.2.2. Genetic Content of the Y Chromosome
2.2.3. Gene Function of the Y Chromosome
2.3. Specific Particularities of the MSY in Relation to Human Population History
3. Y Haplogroups
4. The Y Chromosome in Genetic Epidemiological Studies of Human Complex Traits
4.1. Gender Differences in Relation to Disease
4.2. Study Design
4.2.1. Genome-Wide Association Studies (GWASs)
4.2.2. Expression Quantitative Trait Loci (eQTLs) and Protein Quantitative Trait Loci (pQTLs)
4.3. Use of Y Haplogroups in Genetic Association Studies of Common Complex Traits
4.3.1. Genetic Complexity Inherent to Y Haplogroups
4.3.2. Statistical Power Issues in Studies Involving Y Haplogroups
4.3.3. Population Structure Particularities Specific to Y Haplogroups
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Human Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Painter, T.S. The y-chromosome in mammals. Science 1921, 53, 503–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova, M.; Leroy, P.; Boucekkine, C.; Weissenbach, J.; Bishop, C.; Fellous, M.; Purrello, M.; Fiori, G.; Siniscalco, M. A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 1985, 230, 1403–1406. [Google Scholar] [CrossRef] [PubMed]
- Disteche, C.M.; Casanova, M.; Saal, H.; Friedman, C.; Sybert, V.; Graham, J.; Thuline, H.; Page, D.C.; Fellous, M. Small deletions of the short arm of the Y chromosome in 46,XY females. Proc. Natl. Acad. Sci. USA 1986, 83, 7841–7844. [Google Scholar] [CrossRef] [Green Version]
- Lucotte, G.; Guérin, P.; Hallé, L.; Loirat, F.; Hazout, S. Y chromosome DNA polymorphisms in two African populations. Am. J. Human Genet. 1989, 45, 16–20. [Google Scholar]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ely, D.L.; Turner, M.E. Hypertension in the spontaneously hypertensive rat is linked to the Y chromosome. Hypertension 1990, 16, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef]
- Jobling, M.A.; Pandya, A.; Tyler-Smith, C. The Y chromosome in forensic analysis and paternity testing. Int. J. Legal Med. 1997, 110, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Underhill, P.A.; Shen, P.; Lin, A.A.; Jin, L.; Passarino, G.; Yang, W.H.; Kauffman, E.; Bonné-Tamir, B.; Bertranpetit, J.; Francalacci, P.; et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000, 26, 358–361. [Google Scholar] [CrossRef]
- Ellis, J.A.; Stebbing, M.; Harrap, S.B. Association of the human Y chromosome with high blood pressure in the general population. Hypertension 2000, 36, 731–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charchar, F.J.; Tomaszewski, M.; Padmanabhan, S.; Lacka, B.; Upton, M.N.; Inglis, G.C.; Anderson, N.H.; McConnachie, A.; Zukowska-Szczechowska, E.; Grzeszczak, W.; et al. The Y chromosome effect on blood pressure in two European populations. Hypertension 2002, 39, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Charchar, F.J.; Tomaszewski, M.; Lacka, B.; Zakrzewski, J.; Zukowska-Szczechowska, E.; Grzeszczak, W.; Dominiczak, A.F. Association of the human Y chromosome with cholesterol levels in the general population. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 308–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charchar, F.J.; Bloomer, L.D.; Barnes, T.A.; Cowley, M.J.; Nelson, C.P.; Wang, Y.; Denniff, M.; Debiec, R.; Christofidou, P.; Nankervis, S.; et al. Inheritance of coronary artery disease in men: An analysis of the role of the Y chromosome. Lancet 2012, 379, 915–922. [Google Scholar] [CrossRef] [Green Version]
- Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002, 12, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef]
- Haitjema, S.; van Setten, J.; Eales, J.; van der Laan, S.W.; Gandin, I.; de Vries, J.P.; de Borst, G.J.; Pasterkamp, G.; Asselbergs, F.W.; Charchar, F.J.; et al. Genetic variation within the Y chromosome is not associated with histological characteristics of the atherosclerotic carotid artery or aneurysmal wall. Atherosclerosis 2017, 259, 114–119. [Google Scholar] [CrossRef]
- Kostrzewa, G.; Broda, G.; Konarzewska, M.; Krajewki, P.; Płoski, R. Genetic polymorphism of human Y chromosome and risk factors for cardiovascular diseases: A study in WOBASZ cohort. PLoS ONE 2013, 8, e68155. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, S.; Chen, X.H.; Miller, G.J.; Day, I.N. Non-recombining chromosome Y haplogroups and centromeric HindIII RFLP in relation to blood pressure in 2743 middle-aged Caucasian men from the UK. Human Genet. 2005, 116, 311–318. [Google Scholar] [CrossRef]
- Russo, P.; Venezia, A.; Lauria, F.; Strazzullo, P.; Cappuccio, F.P.; Iacoviello, L.; Barba, G.; Siani, A. HindIII(+/-) polymorphism of the Y chromosome, blood pressure, and serum lipids: No evidence of association in three white populations. Am. J. Hypertens 2006, 19, 331–338. [Google Scholar] [CrossRef]
- Lu, C.; Wen, Y.; Hu, W.; Lu, F.; Qin, Y.; Wang, Y.; Li, S.; Yang, S.; Lin, Y.; Wang, C.; et al. Y chromosome haplogroups based genome-wide association study pinpoints revelation for interactions on non-obstructive azoospermia. Sci. Rep. 2016, 6, 33363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sezgin, E.; Lind, J.M.; Shrestha, S.; Hendrickson, S.; Goedert, J.J.; Donfield, S.; Kirk, G.D.; Phair, J.P.; Troyer, J.L.; O’Brien, S.J.; et al. Association of Y chromosome haplogroup I with HIV progression, and HAART outcome. Human Genet. 2009, 125, 281–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, L.J.; Erzurumluoglu, A.M.; Davey Smith, G.; Rodriguez, S.; Stergiakouli, E. Y Chromosome, Mitochondrial DNA and Childhood Behavioural Traits. Sci. Rep. 2017, 7, 11655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Fernandez Banet, J.; Billis, K.; García Girón, C.; Hourlier, T.; et al. The Ensembl gene annotation system. Database (Oxford) 2016, 2016. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Graves, T.; Fulton, R.S.; Dugan, S.; Ding, Y.; Buhay, C.J.; Kremitzki, C.; et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 2012, 483, 82–86. [Google Scholar] [CrossRef] [Green Version]
- Rozen, S.; Skaletsky, H.; Marszalek, J.D.; Minx, P.J.; Cordum, H.S.; Waterston, R.H.; Wilson, R.K.; Page, D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 2003, 423, 873–876. [Google Scholar] [CrossRef]
- Cruciani, F.; Trombetta, B.; Macaulay, V.; Scozzari, R. About the X-to-Y gene conversion rate. Am. J. Human Genet. 2010, 86, 495–497; author reply 497–498. [Google Scholar] [CrossRef] [Green Version]
- Trombetta, B.; Cruciani, F. Y chromosome palindromes and gene conversion. Human Genet. 2017, 136, 605–619. [Google Scholar] [CrossRef]
- Aitken, R.J.; Marshall Graves, J.A. The future of sex. Nature 2002, 415, 963. [Google Scholar] [CrossRef]
- Hawley, R.S. The human Y chromosome: Rumors of its death have been greatly exaggerated. Cell 2003, 113, 825–828. [Google Scholar] [CrossRef] [Green Version]
- Jobling, M.; Hollox, E.; Hurles, M.; Kivisild, T.; Tyler-Smith, C. Human Evolutionary Genetics, 2nd ed.; Garland Science: New York, NY, USA, 2013. [Google Scholar]
- Graves, J.A. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall Graves, J.A. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev. Genet. 2008, 42, 565–586. [Google Scholar] [CrossRef]
- Arakawa, Y.; Nishida-Umehara, C.; Matsuda, Y.; Sutou, S.; Suzuki, H. X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet. Genome Res. 2002, 99, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Skaletsky, H.; Pyntikova, T.; Minx, P.J.; Graves, T.; Rozen, S.; Wilson, R.K.; Page, D.C. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature 2005, 437, 100–103. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos Trans. R Soc. Lond B Biol. Sci. 2000, 355, 1563–1572. [Google Scholar] [CrossRef] [Green Version]
- Skov, L.; Schierup, M.H.; Consortium, D.P.G. Analysis of 62 hybrid assembled human Y chromosomes exposes rapid structural changes and high rates of gene conversion. PLoS Genet. 2017, 13, e1006834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef] [Green Version]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Harrow, J.; Harte, R.A.; Wallin, C.; Diekhans, M.; Maglott, D.R.; Searle, S.; Farrell, C.M.; Loveland, J.E.; Ruef, B.J.; et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009, 19, 1316–1323. [Google Scholar] [CrossRef] [Green Version]
- Gaudet, P.; Michel, P.A.; Zahn-Zabal, M.; Britan, A.; Cusin, I.; Domagalski, M.; Duek, P.D.; Gateau, A.; Gleizes, A.; Hinard, V.; et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017, 45, D177–D182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girardi, S.K.; Mielnik, A.; Schlegel, P.N. Submicroscopic deletions in the Y chromosome of infertile men. Human Reprod. 1997, 12, 1635–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, P.H.; Edelmann, A.; Kirsch, S.; Henegariu, O.; Hirschmann, P.; Kiesewetter, F.; Köhn, F.M.; Schill, W.B.; Farah, S.; Ramos, C.; et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Human Mol. Genet. 1996, 5, 933–943. [Google Scholar] [CrossRef]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef]
- Meyfour, A.; Pooyan, P.; Pahlavan, S.; Rezaei-Tavirani, M.; Gourabi, H.; Baharvand, H.; Salekdeh, G.H. Chromosome-Centric Human Proteome Project Allies with Developmental Biology: A Case Study of the Role of Y Chromosome Genes in Organ Development. J. Proteome Res. 2017, 16, 4259–4272. [Google Scholar] [CrossRef]
- Underhill, P.A.; Kivisild, T. Use of y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev. Genet. 2007, 41, 539–564. [Google Scholar] [CrossRef] [Green Version]
- Hammer, M.F.; Spurdle, A.B.; Karafet, T.; Bonner, M.R.; Wood, E.T.; Novelletto, A.; Malaspina, P.; Mitchell, R.J.; Horai, S.; Jenkins, T.; et al. The geographic distribution of human Y chromosome variation. Genetics 1997, 145, 787–805. [Google Scholar]
- Watanabe, Y.; Naka, I.; Khor, S.S.; Sawai, H.; Hitomi, Y.; Tokunaga, K.; Ohashi, J. Analysis of whole Y-chromosome sequences reveals the Japanese population history in the Jomon period. Sci. Rep. 2019, 9, 8556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupanloup, I.; Pereira, L.; Bertorelle, G.; Calafell, F.; Prata, M.J.; Amorim, A.; Barbujani, G. A recent shift from polygyny to monogamy in humans is suggested by the analysis of worldwide Y-chromosome diversity. J. Mol. Evol. 2003, 57, 85–97. [Google Scholar] [CrossRef]
- Hammer, M.F.; Redd, A.J.; Wood, E.T.; Bonner, M.R.; Jarjanazi, H.; Karafet, T.; Santachiara-Benerecetti, S.; Oppenheim, A.; Jobling, M.A.; Jenkins, T.; et al. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc. Natl. Acad. Sci. USA 2000, 97, 6769–6774. [Google Scholar] [CrossRef] [Green Version]
- Karmin, M.; Saag, L.; Vicente, M.; Wilson Sayres, M.A.; Järve, M.; Talas, U.G.; Rootsi, S.; Ilumäe, A.M.; Mägi, R.; Mitt, M.; et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015, 25, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, S. The Distribution of Gene Frequencies in Populations of Polyploids. Proc. Natl. Acad. Sci. USA 1938, 24, 372–377. [Google Scholar] [CrossRef] [Green Version]
- Jobling, M.A.; Tyler-Smith, C. The human Y chromosome: An evolutionary marker comes of age. Nat. Rev. Genet. 2003, 4, 598–612. [Google Scholar] [CrossRef]
- Giles, R.E.; Blanc, H.; Cann, H.M.; Wallace, D.C. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 6715–6719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, M.; Sato, K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta 2013, 1833, 1979–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinnery, P.F. Inheritance of mitochondrial disorders. Mitochondrion 2002, 2, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Cavalli-Sforza, L.L.; Feldman, M.W. The application of molecular genetic approaches to the study of human evolution. Nat. Genet. 2003, 33, 266–275. [Google Scholar] [CrossRef]
- Seielstad, M.T.; Minch, E.; Cavalli-Sforza, L.L. Genetic evidence for a higher female migration rate in humans. Nat. Genet. 1998, 20, 278–280. [Google Scholar] [CrossRef]
- Bolnick, D.A.; Bolnick, D.I.; Smith, D.G. Asymmetric male and female genetic histories among Native Americans from Eastern North America. Mol. Biol. Evol. 2006, 23, 2161–2174. [Google Scholar] [CrossRef] [Green Version]
- Oota, H.; Settheetham-Ishida, W.; Tiwawech, D.; Ishida, T.; Stoneking, M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat. Genet. 2001, 29, 20–21. [Google Scholar] [CrossRef]
- Lippold, S.; Xu, H.; Ko, A.; Li, M.; Renaud, G.; Butthof, A.; Schröder, R.; Stoneking, M. Human paternal and maternal demographic histories: Insights from high-resolution Y chromosome and mtDNA sequences. Investig. Genet. 2014, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Maan, A.A.; Eales, J.; Akbarov, A.; Rowland, J.; Xu, X.; Jobling, M.A.; Charchar, F.J.; Tomaszewski, M. The Y chromosome: A blueprint for men’s health? Eur. J. Hum. Genet. 2017, 25, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Seielstad, M.T.; Perez-Lezaun, A.; Feldman, M.W. Population growth of human Y chromosomes: A study of Y chromosome microsatellites. Mol. Biol. Evol. 1999, 16, 1791–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teitz, L.S.; Pyntikova, T.; Skaletsky, H.; Page, D.C. Selection Has Countered High Mutability to Preserve the Ancestral Copy Number of Y Chromosome Amplicons in Diverse Human Lineages. Am. J. Hum. Genet. 2018, 103, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendez, F.L.; Krahn, T.; Schrack, B.; Krahn, A.M.; Veeramah, K.R.; Woerner, A.E.; Fomine, F.L.; Bradman, N.; Thomas, M.G.; Karafet, T.M.; et al. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am. J. Hum. Genet. 2013, 92, 454–459. [Google Scholar] [CrossRef] [Green Version]
- Elhaik, E.; Tatarinova, T.V.; Klyosov, A.A.; Graur, D. The ‘extremely ancient’ chromosome that isn’t: A forensic bioinformatic investigation of Albert Perry’s X-degenerate portion of the Y chromosome. Eur. J. Human Genet. 2014, 22, 1111–1116. [Google Scholar] [CrossRef] [Green Version]
- Mendez, F.L.; Veeramah, K.R.; Thomas, M.G.; Karafet, T.M.; Hammer, M.F. Reply to ‘The ‘extremely ancient’ chromosome that isn‘t’ by Elhaik et al. Eur. J. Human Genet. 2015, 23, 564–567. [Google Scholar] [CrossRef] [Green Version]
- Elhaik, E.; Tatarinova, T.V.; Klyosov, A.A.; Graur, D. Reply to Mendez et al: The ‘extremely ancient’ chromosome that still isn’t. Eur J. Human Genet. 2015, 23, 567–568. [Google Scholar] [CrossRef] [Green Version]
- Karafet, T.M.; Mendez, F.L.; Meilerman, M.B.; Underhill, P.A.; Zegura, S.L.; Hammer, M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008, 18, 830–838. [Google Scholar] [CrossRef] [Green Version]
- Calafell, F.; Larmuseau, M.H.D. The Y chromosome as the most popular marker in genetic genealogy benefits interdisciplinary research. Human Genet. 2017, 136, 559–573. [Google Scholar] [CrossRef]
- Kayser, M. Forensic use of Y-chromosome DNA: A general overview. Human Genet. 2017, 136, 621–635. [Google Scholar] [CrossRef] [Green Version]
- Underhill, P.A.; Jin, L.; Zemans, R.; Oefner, P.J.; Cavalli-Sforza, L.L. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc. Natl. Acad. Sci. USA 1996, 93, 196–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underhill, P.A.; Jin, L.; Lin, A.A.; Mehdi, S.Q.; Jenkins, T.; Vollrath, D.; Davis, R.W.; Cavalli-Sforza, L.L.; Oefner, P.J. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997, 7, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- de Knijff, P.; Kayser, M.; Caglià, A.; Corach, D.; Fretwell, N.; Gehrig, C.; Graziosi, G.; Heidorn, F.; Herrmann, S.; Herzog, B.; et al. Chromosome Y microsatellites: Population genetic and evolutionary aspects. Int. J. Legal Med. 1997, 110, 134–149. [Google Scholar] [CrossRef]
- Jobling, M.A.; Bouzekri, N.; Taylor, P.G. Hypervariable digital DNA codes for human paternal lineages: MVR-PCR at the Y-specific minisatellite, MSY1 (DYF155S1). Human Mol. Genet. 1998, 7, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Seielstad, M.; Bekele, E.; Ibrahim, M.; Touré, A.; Traoré, M. A view of modern human origins from Y chromosome microsatellite variation. Genome Res. 1999, 9, 558–567. [Google Scholar]
- Wells, R.S.; Yuldasheva, N.; Ruzibakiev, R.; Underhill, P.A.; Evseeva, I.; Blue-Smith, J.; Jin, L.; Su, B.; Pitchappan, R.; Shanmugalakshmi, S.; et al. The Eurasian heartland: A continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. USA 2001, 98, 10244–10249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, S.D.; Santos, F.R.; Bianchi, N.O.; Bravi, C.M.; Carnese, F.R.; Rothhammer, F.; Gerelsaikhan, T.; Munkhtuja, B.; Oyunsuren, T. A major founder Y-chromosome haplotype in Amerindians. Nat. Genet. 1995, 11, 15–16. [Google Scholar] [CrossRef]
- Hammer, M.F.; Karafet, T.; Rasanayagam, A.; Wood, E.T.; Altheide, T.K.; Jenkins, T.; Griffiths, R.C.; Templeton, A.R.; Zegura, S.L. Out of Africa and back again: Nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 1998, 15, 427–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roewer, L.; Croucher, P.J.; Willuweit, S.; Lu, T.T.; Kayser, M.; Lessig, R.; de Knijff, P.; Jobling, M.A.; Tyler-Smith, C.; Krawczak, M. Signature of recent historical events in the European Y-chromosomal STR haplotype distribution. Human Genet. 2005, 116, 279–291. [Google Scholar] [CrossRef]
- Chen, X.H.; Rodríguez, S.; Hawe, E.; Talmud, P.J.; Miller, G.J.; Underhill, P.; Humphries, S.E.; Day, I.N. Evidence of admixture from haplotyping in an epidemiological study of UK Caucasian males: Implications for association analyses. Human Hered 2004, 57, 142–155. [Google Scholar] [CrossRef]
- Rootsi, S.; Behar, D.M.; Järve, M.; Lin, A.A.; Myres, N.M.; Passarelli, B.; Poznik, G.D.; Tzur, S.; Sahakyan, H.; Pathak, A.K.; et al. Phylogenetic applications of whole Y-chromosome sequences and the Near Eastern origin of Ashkenazi Levites. Nat. Commun. 2013, 4, 2928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayser, M.; Lao, O.; Anslinger, K.; Augustin, C.; Bargel, G.; Edelmann, J.; Elias, S.; Heinrich, M.; Henke, J.; Henke, L.; et al. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Human Genet. 2005, 117, 428–443. [Google Scholar] [CrossRef]
- Lao, O.; Lu, T.T.; Nothnagel, M.; Junge, O.; Freitag-Wolf, S.; Caliebe, A.; Balascakova, M.; Bertranpetit, J.; Bindoff, L.A.; Comas, D.; et al. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008, 18, 1241–1248. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.; Khalifa, A.O.; Isali, I.; Shukla, S. Prostate cancer susceptibility and growth linked to Y chromosome genes. Front. Biosci. (Elite Ed.) 2018, 10, 423–436. [Google Scholar] [CrossRef] [Green Version]
- Kido, T.; Lau, Y.F. Roles of the Y chromosome genes in human cancers. Asian J. Androl 2015, 17, 373–380. [Google Scholar] [CrossRef]
- Lleo, A.; Oertelt-Prigione, S.; Bianchi, I.; Caliari, L.; Finelli, P.; Miozzo, M.; Lazzari, R.; Floreani, A.; Donato, F.; Colombo, M.; et al. Y chromosome loss in male patients with primary biliary cirrhosis. J. Autoimmun. 2013, 41, 87–91. [Google Scholar] [CrossRef]
- Persani, L.; Bonomi, M.; Lleo, A.; Pasini, S.; Civardi, F.; Bianchi, I.; Campi, I.; Finelli, P.; Miozzo, M.; Castronovo, C.; et al. Increased loss of the Y chromosome in peripheral blood cells in male patients with autoimmune thyroiditis. J. Autoimmun. 2012, 38, J193–J196. [Google Scholar] [CrossRef]
- Khan, S.I.; Andrews, K.L.; Jennings, G.L.; Sampson, A.K.; Chin-Dusting, J.P.F. Y Chromosome, Hypertension and Cardiovascular Disease: Is Inflammation the Answer? Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Institute of Medicine (US) Committee on Understanding the Biologhy of Sex and Gender Differences; Wizemann, T.M.; Pardue, M.L. Exploring the Biological Contributions to Human Health: Does Sex Matter? The National Acadamies Press (US): Washington, DC, USA, 2001. [Google Scholar]
- McGrath, J.; Saha, S.; Chant, D.; Welham, J. Schizophrenia: A concise overview of incidence, prevalence, and mortality. Epidemiol. Rev. 2008, 30, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Després, J.P.; Fullerton, H.J.; et al. Executive Summary: Heart Disease and Stroke Statistics—2016 Update: A Report From the American Heart Association. Circulation 2016, 133, 447–454. [Google Scholar] [CrossRef]
- Somers, E.C.; Thomas, S.L.; Smeeth, L.; Schoonen, W.M.; Hall, A.J. Incidence of systemic lupus erythematosus in the United Kingdom, 1990–1999. Arthritis Care Rheum. 2007, 57, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Steyn, F.J.; McCombe, P.A. Gender differences in autoimmune disease. Front. Neuroendocrinol. 2014, 35, 347–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitacre, C.C.; Reingold, S.C.; O’Looney, P.A. A gender gap in autoimmunity. Science 1999, 283, 1277–1278. [Google Scholar] [CrossRef]
- Voskuhl, R.R.; Sawalha, A.H.; Itoh, Y. Sex chromosome contributions to sex differences in multiple sclerosis susceptibility and progression. Mult. Scler. 2018, 24, 22–31. [Google Scholar] [CrossRef]
- Anand, S.S.; Islam, S.; Rosengren, A.; Franzosi, M.G.; Steyn, K.; Yusufali, A.H.; Keltai, M.; Diaz, R.; Rangarajan, S.; Yusuf, S.; et al. Risk factors for myocardial infarction in women and men: Insights from the INTERHEART study. Eur. Heart J. 2008, 29, 932–940. [Google Scholar] [CrossRef] [Green Version]
- Bao, A.M.; Swaab, D.F. Sexual differentiation of the human brain: Relation to gender identity, sexual orientation and neuropsychiatric disorders. Front. Neuroendocrinol. 2011, 32, 214–226. [Google Scholar] [CrossRef]
- Ding, E.L.; Song, Y.; Malik, V.S.; Liu, S. Sex differences of endogenous sex hormones and risk of type 2 diabetes: A systematic review and meta-analysis. JAMA 2006, 295, 1288–1299. [Google Scholar] [CrossRef]
- Adams Waldorf, K.M.; Nelson, J.L. Autoimmune disease during pregnancy and the microchimerism legacy of pregnancy. Immunol. Invest. 2008, 37, 631–644. [Google Scholar] [CrossRef]
- Terwilliger, J.D.; Göring, H.H. Gene mapping in the 20th and 21st centuries: Statistical methods, data analysis, and experimental design. 2000. Human Biol. 2009, 81, 663–728. [Google Scholar] [CrossRef] [PubMed]
- García, E.C.; González, P.; Castro, M.G.; Alvarez, R.; Reguero, J.R.; Batalla, A.; Cortina, A.; Alvarez, V. Association between genetic variation in the Y chromosome and hypertension in myocardial infarction patients. Am. J. Med. Genet. A 2003, 122A, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Suto, J.; Satou, K. Effect of the Y chromosome on plasma high-density lipoprotein-cholesterol levels in Y-chromosome-consomic mouse strains. BMC Res. Notes 2014, 7, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kren, V.; Qi, N.; Krenova, D.; Zidek, V.; Sladká, M.; Jáchymová, M.; Míková, B.; Horky, K.; Bonne, A.; Van Lith, H.A.; et al. Y-chromosome transfer induces changes in blood pressure and blood lipids in SHR. Hypertension 2001, 37, 1147–1152. [Google Scholar] [CrossRef] [Green Version]
- Dickey, C.; Toot, J.; Terwilliger, M.; Payne, R.; Turner, M.; Ely, D. The SHR Y chromosome increases cardiovascular, endocrine, and behavioral responses to stress compared to the WKY Y chromosome. Physiol. Behav. 2012, 106, 101–108. [Google Scholar] [CrossRef]
- Prokop, J.W.; Tsaih, S.W.; Faber, A.B.; Boehme, S.; Underwood, A.C.; Troyer, S.; Playl, L.; Milsted, A.; Turner, M.E.; Ely, D.; et al. The phenotypic impact of the male-specific region of chromosome-Y in inbred mating: The role of genetic variants and gene duplications in multiple inbred rat strains. Biol. Sex. Differ. 2016, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voskarides, K.; Hadjipanagi, D.; Papazachariou, L.; Griffin, M.; Panayiotou, A.G. Evidence for contribution of the y chromosome in atherosclerotic plaque occurrence in men. Genet. Test. Mol. Biomarkers 2014, 18, 552–556. [Google Scholar] [CrossRef]
- Bloomer, L.D.; Nelson, C.P.; Eales, J.; Denniff, M.; Christofidou, P.; Debiec, R.; Moore, J.; Zukowska-Szczechowska, E.; Goodall, A.H.; Thompson, J.; et al. Male-specific region of the Y chromosome and cardiovascular risk: Phylogenetic analysis and gene expression studies. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1722–1727. [Google Scholar] [CrossRef] [Green Version]
- O’Keeffe, L.M.; Howe, L.D.; Fraser, A.; Hughes, A.D.; Wade, K.H.; Anderson, E.L.; Lawlor, D.A.; Erzurumluoglu, A.M.; Davey-Smith, G.; Rodriguez, S.; et al. Associations of Y chromosomal haplogroups with cardiometabolic risk factors and subclinical vascular measures in males during childhood and adolescence. Atherosclerosis 2018, 274, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Shinka, T.; Nozawa, S.; Yoshiike, M.; Koh, E.; Kanaya, J.; Namiki, M.; Matsumiya, K.; Tsujimura, A.; Komatsu, K.; et al. Y chromosome haplogroup D2a1 is significantly associated with high levels of luteinizing hormone in Japanese men. Andrology 2015, 3, 520–525. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.; Cañadas-Garre, M.; Chambers, R.; Maxwell, A.P.; McKnight, A.J. The Challenges of Chromosome Y Analysis and the Implications for Chronic Kidney Disease. Front. Genet. 2019, 10, 781. [Google Scholar] [CrossRef]
- Winham, S.J.; de Andrade, M.; Miller, V.M. Genetics of cardiovascular disease: Importance of sex and ethnicity. Atherosclerosis 2015, 241, 219–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B.; GTEx Consortium; Laboratory, Data Analysis & Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Erzurumluoglu, A.M.; Elsworth, B.L.; Kemp, J.P.; Howe, L.; Haycock, P.C.; Hemani, G.; Tansey, K.; Laurin, C.; Pourcain, B.S.; et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017, 33, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Riley, V.; Erzurumluoglu, A.M.; Rodriguez, S.; Bonilla, C. Mitochondrial DNA Haplogroups and Breast Cancer Risk Factors in the Avon Longitudinal Study of Parents and Children (ALSPAC). Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Lawson, D.J.; Davies, N.M.; Haworth, S.; Ashraf, B.; Howe, L.; Crawford, A.; Hemani, G.; Davey Smith, G.; Timpson, N.J. Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity? Human Genet. 2020, 139, 23–41. [Google Scholar] [CrossRef] [Green Version]
- Erzurumluoglu, A.M.; Baird, D.; Richardson, T.G.; Timpson, N.J.; Rodriguez, S. Using Y-Chromosomal Haplogroups in Genetic Association Studies and Suggested Implications. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Makowsky, R.; Yan, Q.; Wiener, H.W.; Sandel, M.; Aissani, B.; Tiwari, H.K.; Shrestha, S. The utility of mitochondrial and y chromosome phylogenetic data to improve correction for population stratification. Front. Genet. 2012, 3, 301. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Shakhbazov, K.; Westra, H.J.; Esko, T.; Henders, A.K.; McRae, A.F.; Yang, J.; Gibson, G.; Martin, N.G.; Metspalu, A.; et al. Detection and replication of epistasis influencing transcription in humans. Nature 2014, 508, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Hunt, S.E.; McLaren, W.; Gil, L.; Thormann, A.; Schuilenburg, H.; Sheppard, D.; Parton, A.; Armean, I.M.; Trevanion, S.J.; Flicek, P.; et al. Ensembl variation resources. Database (Oxford) 2018, 2018. [Google Scholar] [CrossRef]
- Ramasamy, A.; Trabzuni, D.; Guelfi, S.; Varghese, V.; Smith, C.; Walker, R.; De, T.; Coin, L.; de Silva, R.; Cookson, M.R.; et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014, 17, 1418–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlen, M.; Bandrowski, A.; Carr, S.; Edwards, A.; Ellenberg, J.; Lundberg, E.; Rimm, D.L.; Rodriguez, H.; Hiltke, T.; Snyder, M.; et al. A proposal for validation of antibodies. Nat. Methods 2016, 13, 823–827. [Google Scholar] [CrossRef]
- Speliotes, E.K.; Willer, C.J.; Berndt, S.I.; Monda, K.L.; Thorleifsson, G.; Jackson, A.U.; Lango Allen, H.; Lindgren, C.M.; Luan, J.; Magi, R.; et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010, 42, 937–948. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.G. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 2004, 27, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, C.C.; Yang, C.; Meng, H.; Agbagwa, I.O.; Wang, L.X.; Wang, Y.; Yan, S.; Ren, S.; Sun, Y.; et al. Epigenetic Pattern on the Human Y Chromosome Is Evolutionarily Conserved. PLoS ONE 2016, 11, e0146402. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Year | Description | References |
|---|---|---|
| 1985 | ChrY utilised for evolutionary studies ‘genetic distance’ genealogy | [4] |
| 1986 | First evidence that the male-determining region is located on the short arm of the Y chromosome | [5] |
| 1989 | ChrY polymorphisms utilised in phylogenetics | [6] |
| 1990 | First gene, sex-determining region Y (SRY), mapped onto ChrY | [7] |
| 1990 | Animal models used to investigate the influence of ChrY on hypertension | [8] |
| 1991 | Sry used in animal models to form transgenic mice | [9] |
| 1997 | ChrY utilised for forensic science and paternity testing | [10] |
| 2000 | ChrY utilised for phylogenetics | [11] |
| 2000 onwards | Association between hind III restriction fragment polymorphism and cardiovascular disease detected | [12,13,14,15] |
| 2002 | Y-chromosomal haplogroups established | [16] |
| 2003 | MSY first sequenced | [17] |
| 2005 onwards | Unconvincing evidence relating the MSY to cardiovascular disease risk is reported | [18,19,20,21] |
| 2009 onwards | Y haplotypes utilised to investigate association between ChrY and other complex traits | [22,23,24] |
| 2016 | Exploration of the function of genes identified on within MSY | [25] |
| Study Design | Basis | Strengths | Limitations | Reference |
|---|---|---|---|---|
| Linkage analysis | First-degree relatives are compared in order to ascertain the potential for a genetic component of disease susceptibility | 1- Successful in the identification of highly penetrant genetic variants related to Mendelian traits or monogenic disorders | 1- Limited application for complex traits due to the use of individuals that share similar genetic and environmental constituents, establishing true effects in multifactorial traits is limited | [44] |
| Candidate gene studies | A particular gene is studied based on biological plausibility. Variation at this gene is investigated in genetic association studies | 1- Highly specific for genetic variation which focuses on the MSY | 1- Locus selected in absence of understanding of its function and potential effects 2- Highly vulnerable to chance making conflicting evidence more likely | [12,13,14,19,20,21,103] |
| Animal models | Animal studies are used as a framework for looking at human disease | 1- Allows careful control and manipulation of both genetic and external environment to isolate the effects of the MSY | 1- Application of animal models to human disease makes two key assumptions that may be incorrect:
| [104,105,106,107] |
| GWAS | Case-control study design used to look for common genetic variants more frequently identified in those with particular diseases | 1- Allows genotype-first analysis of the MSY for which understanding of genetic content is limited | 1- Sex chromosomes are routinely excluded from these study types as the entirety of the MSY is effectively in linkage disequilibrium 2- Haplotypes and haplogroups used as a basis for this type of analysis may be flawed 3- Frequency of haplotypes within haplogroups in different cohorts may be sufficiently different to dilute or exaggerate relationships seen in other populations | [15,18,22,23,108,109,110,111] |
| GWAS Features | Y Chromosome vs. Autosomes | Implication | Possible Solution(s) | References |
|---|---|---|---|---|
| Statistical power | Only men inherit a Y chromosome | ~50% reduction in sample size | 1- Include the Y chromosome in GWASs—could be mandated by funders 2- Share GWAS summary results (e.g., via LD Hub, GWAS Catalogue) 3- Increase sample sizes | [2,112,115] |
| LD structure | All the common variants in the MSY are in LD | Identifying the causal variant is very difficult | 1- Larger sample sizes 2- Fine mapping by (i) sequencing the MSY and the associated region, (ii) carrying out transethnic studies and/or (iii) Y-DNA haplogroup association studies 3- Functional analyses (e.g., single SNP editing) | [15,24,116] |
| Population stratification | Principal components calculated using autosomal SNPs are not applicable | Potential overadjustment and loss of statistical power | 1- Sensitivity analyses with and without genetic principal components 2- Sensitivity analyses with and without Y-DNA haplogroup information as a covariate in (male-only) GWASs and looking for SNPs with significant differences in effect sizes | [117,118,119] |
| Complex loci | Many highly variable regions and repetitive sequences | Variant calling may not be accurate | Only include variants called with high confidence | [17] |
| Colocalisation of eQTL and GWAS signals | No eQTLs identified for the Y chromosome | GWAS-eQTL colocalisation analysis cannot be carried out at present | Initiate trans-ethnic eeGWASs and/or study rare variants on the Y chromosome | [114] |
| Pleiotropy | No conclusive GWAS signal identified | Not much information to link potential findings with other biological pathways | 1- Initiate a consortium to identify associations on the MSY 2- Carry out PheWASs using all Y chromosomal SNPs (incl. rare variants) | [2] |
| Gene–gene interactions | Very few examples identified in autosomes. None with SNPs on the Y chromosome | Almost no statistical power to detect small effects | Hypothesis driven approaches (e.g., between SNPs associated with obesity/CVD and SNPs in/near UTY—a gene expressed in non-gonadal tissues) | [120] |
| Gene/protein Expression | Many MSY genes/proteins are not expressed at detectable levels in non-gonadal tissues | Identified associations are likely to be biologically implausible if not expressed in disease-relevant tissue | Query the Human Protein Atlas to check whether the putatively causal gene/protein is active in a relevant tissue (Figure 4) | [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, K.; Erzurumluoglu, A.M.; Rodriguez, S. The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits. Genes 2020, 11, 1273. https://doi.org/10.3390/genes11111273
Parker K, Erzurumluoglu AM, Rodriguez S. The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits. Genes. 2020; 11(11):1273. https://doi.org/10.3390/genes11111273
Chicago/Turabian StyleParker, Katherine, A. Mesut Erzurumluoglu, and Santiago Rodriguez. 2020. "The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits" Genes 11, no. 11: 1273. https://doi.org/10.3390/genes11111273
APA StyleParker, K., Erzurumluoglu, A. M., & Rodriguez, S. (2020). The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits. Genes, 11(11), 1273. https://doi.org/10.3390/genes11111273






