Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction, Genotyping and SNP Calling
2.3. GWAS of the Carioca Subset
2.4. Statistical Analysis
2.5. Andean Introgression into the Mesoamerican Cultivars
2.6. Genome-Wide Association Studies
3. Results
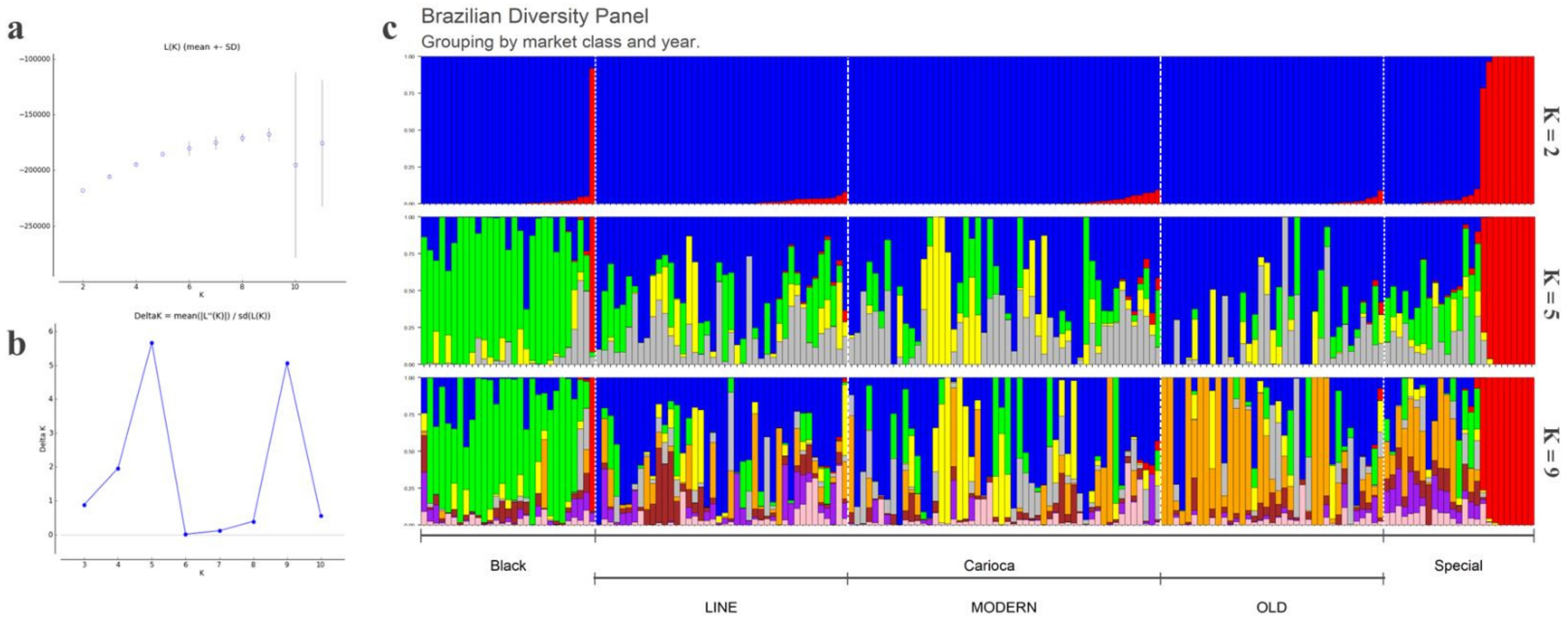
3.1. Population Structure, General SNP Diversity, and Genetic Relationships
3.2. Genetic Relationships and Diversity
3.3. Differentiation among the Commercial Classes
3.4. Diversity Gain of the Carioca Variety
3.5. Linkage Disequilibrium Decay
3.6. Andean Introgression
3.7. Genome Wide Association Studies
4. Discussion
4.1. Andean Introgression into Brazilian Mesoamerican Cultivars
4.2. Diversity, Genetic Structuring, and Differentiation in Commercial Classes
4.3. Gain in Genetic Diversity after Almost Half a Century of Genetic Breeding
4.4. Validation of the Carioca Diversity Subset in Genome Wide Association Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cortinovis, G.; Di Vittori, V.; Bellucci, E.; Bitocchi, E.; Papa, R. Adaptation to novel environments during crop diversification. Curr. Opin. Plant Biol. 2020, 56, 203–217. [Google Scholar] [CrossRef]
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)—Model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar]
- Bellucci, E.; Bitocchi, E.; Rau, D.; Rodriguez, M.; Biagetti, E.; Giardini, A.; Attene, G.; Nanni, L.; Papa, R. Genomics of origin, domestication and evolution of Phaseolus vulgaris. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; ISBN 9789400775725. [Google Scholar]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Pires, C.V.; Oliveira, M.G.A.; Cruz, G.A.D.R.; Mendes, F.Q.; De Rezende, S.T.; Moreira, M.A. Composição físico-química de diferentes cultivares de feijão (Phaseolus vulgaris L.). Alimentos e Nutricao Araraquara 2005, 16, 157–162. [Google Scholar]
- FAOSTAT Food and Agriculture Organization of the United Nations (FAO). Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 6 July 2020).
- CONAB Acompanhamento da Safra Brasileira de Grãos: Safra 2018/19 - Décimo Segundo Levantamento. Available online: https://www.conab.gov.br/info-agro/safras/graos/boletim-da-safra-de-graos/ (accessed on 10 June 2020).
- Bitocchi, E.; Nanni, L.; Bellucci, E.; Rossi, M.; Giardini, A.; Zeuli, P.S.; Logozzo, G.; Stougaard, J.; McClean, P.; Attene, G.; et al. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 2012, 109, E788–E796. [Google Scholar] [CrossRef] [Green Version]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef]
- Gepts, P.; Bliss, F.A. Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II. Europe and Africa. Econ. Bot. 1988, 42, 86–104. [Google Scholar] [CrossRef]
- Koinange, E.M.K.; Gepts, P. Hybrid weakness in wild Phaseolus vulgaris L. J. Hered. 1992, 83, 135–139. [Google Scholar] [CrossRef]
- Singh, S.P.; Nodari, R.; Gepts, P. Genetic Diversity in Cultivated Common Bean: I. Allozymes. Crop Sci. 1991, 31, 19–23. [Google Scholar] [CrossRef]
- McClean, P.E.; Lee, R.K.; Miklas, P.N. Sequence diversity analysis of dihydroflavonol 4-reductase intron 1 in common bean. Genome 2004, 47, 266–280. [Google Scholar] [CrossRef]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [Green Version]
- Angioi, S.A.; Rau, D.; Attene, G.; Nanni, L.; Bellucci, E.; Logozzo, G.; Negri, V.; Zeuli, P.L.S.; Papa, R. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theor. Appl. Genet. 2010, 121, 829–843. [Google Scholar] [CrossRef]
- Ariani, A.; Berny Mier y Teran, J.C.; Gepts, P. Genome-wide identification of SNPs and copy number variation in common bean (Phaseolus vulgaris L.) using genotyping-by-sequencing (GBS). Mol. Breed. 2016, 36. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, M.; Rau, D.; Bitocchi, E.; Bellucci, E.; Biagetti, E.; Carboni, A.; Gepts, P.; Nanni, L.; Papa, R.; Attene, G. Landscape genetics, adaptive diversity and population structure in Phaseolus vulgaris. New Phytol. 2016, 209, 1781–1794. [Google Scholar] [CrossRef]
- Cortinovis, G.; Frascarelli, G.; Di Vittori, V.; Papa, R. Current state and perspectives in population genomics of the common bean. Plants 2020, 9, 330. [Google Scholar] [CrossRef] [Green Version]
- Chiorato, A.F.; Carbonell, S.A.M. O Melhoramento Genético do Feijoeiro no Instituto Agronômico IAC (1932 a 2014); O AGRONÔMICO: Campinas, Brazil, 2014. [Google Scholar]
- De Almeida, L.D.; Leitão Filho, H.F.; Miyasaka, S. Características do feijão Carioca, um nôvo cultivar. Bragantia 1971, 30. [Google Scholar] [CrossRef]
- Pereira, H.S.; Mota, A.P.S.; Rodrigues, L.A.; de Souza, T.L.P.O.; Melo, L.C. Genetic diversity among common bean cultivars based on agronomic traits and molecular markers and application to recommendation of parent lines. Euphytica 2019, 215, 1–16. [Google Scholar] [CrossRef]
- Campa, A.; Murube, E.; Ferreira, J.J. Genetic diversity, population structure, and linkage disequilibrium in a spanish common bean diversity panel revealed through genotyping-by-sequencing. Genes 2018, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gioia, T.; Logozzo, G.; Attene, G.; Bellucci, E.; Benedettelli, S.; Negri, V.; Papa, R.; Spagnoletti Zeuli, P. Evidence for Introduction Bottleneck and Extensive Inter-Gene Pool (Mesoamerica x Andes) Hybridization in the European Common Bean (Phaseolus vulgaris L.) Germplasm. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gioia, T.; Logozzo, G.; Marzario, S.; Zeuli, P.S.; Gepts, P. Evolution of SSR diversity from wild types to U.S. Advanced cultivars in the Andean and Mesoamerican domestications of common bean (Phaseolus vulgaris). PLoS ONE 2019, 14, e0211342. [Google Scholar] [CrossRef] [Green Version]
- Cooper, H.D.; Spillane, C.; Hodgkin, T. Broadening the genetic base of crops: an overview. In Broadening the Genetic Base of Crop Production; CAB International: Wallingford, UK, 2009; pp. 1–23. [Google Scholar]
- Elias, H.T.; Vidigal, M.C.G.; Gonela, A.; Vogt, G.A. Variabilidade genética em germoplasma tradicional de feijão-preto em Santa Catarina. Pesquisa Agropecuaria Brasileira 2007, 42, 1443–1449. [Google Scholar] [CrossRef] [Green Version]
- Cargnelutti Filho, A.; Ribeiro, N.D.; Dos Reis, R.C.P.; De Souza, J.R.; Jost, E. Comparação de métodos de agrupamento para o estudo da divergência genética em cultivares de feijão. Ciencia Rural 2008, 38, 2138–2145. [Google Scholar] [CrossRef]
- Cabral, P.D.S.; Soares, T.C.B.; de Passos Lima, A.B.; de Souza Alves, D.; Nunes, J.A. Diversidade genética de acessos de feijão comum por caracteres agronômicos. Revista Ciencia Agronomica 2011, 42, 898–905. [Google Scholar] [CrossRef]
- Delfini, J.; Moda-Cirino, V.; Ruas, C.D.F.; Dos Santos Neto, J.; Ruas, P.M.; Buratto, J.S.; Ruas, E.A.; Gonçalves, L.S.A. Distinctness of Brazilian common bean cultivars with carioca and black grain by means of morphoagronomic and molecular descriptors. PLoS ONe 2017, 12. [Google Scholar] [CrossRef]
- Guidoti, D.T.; Gonela, A.; Vidigal, M.C.G.; Conrado, T.V.; Romani, I. Interrelationship between morphological, agronomic and molecular characteristics in the analysis of common bean genetic diversity. Acta Scientiarum Agronomy 2018, 40, 33032. [Google Scholar] [CrossRef] [Green Version]
- Benchimol, L.L.; De Campos, T.; Carbonell, S.A.M.; Colombo, C.A.; Chioratto, A.F.; Formighieri, E.F.; Gouvêa, L.R.L.; Souza, A.P. De Structure of genetic diversity among common bean (Phaseolus vulgaris L.) varieties of Mesoamerican and Andean origins using new developed microsatellite markers. Genet. Resour. Crop. Evol. 2007, 54, 1747–1762. [Google Scholar] [CrossRef]
- Cortés, A.J.; Chavarro, M.C.; Blair, M.W. SNP marker diversity in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2011, 123, 827–845. [Google Scholar] [CrossRef]
- Perseguini, J.M.K.C.; Chioratto, A.F.; Zucchi, M.I.; Colombo, C.A.; Carbonell, S.A.M.; Mondego, J.M.C.; Gazaffi, R.; Garcia, A.A.F.; de Campos, T.; de Souza, A.P.; et al. Genetic diversity in cultivated carioca common beans based on molecular marker analysis. Genet. Mol. Biol. 2011, 34, 88–102. [Google Scholar] [CrossRef] [Green Version]
- Díaz, L.M.; Buendía, H.F.; Duque, M.C.; Blair, M.W. Genetic diversity of Colombian landraces of common bean as detected through the use of silver-stained and fluorescently labelled microsatellites. Plant. Genet. Resour. Charact. Util. 2011, 9, 86–96. [Google Scholar] [CrossRef]
- Veloso, J.S.; Silva, W.; Pinheiro, L.R.; dos Santos, J.B.; Fonseca, N.S.; Euzebio, M.P. Genetic divergence of common bean cultivars. Genet. Mol. Res. 2015, 14, 11281–11291. [Google Scholar] [CrossRef]
- Burle, M.L.; Fonseca, J.R.; Kami, J.A.; Gepts, P. Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor. Appl. Genet. 2010, 121, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Jia, G.; Hyten, D.L.; Jenkins, J.; Hwang, E.Y.; Schroeder, S.G.; Osorno, J.M.; Schmutz, J.; Jackson, S.A.; McClean, P.E.; et al. SNP assay development for linkage map construction, anchoring whole-genome sequence, and other genetic and genomic applications in common bean. G3 Genes Genomes Genet. 2015, 5, 2285–2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perseguini, J.M.K.C.; Silva, G.M.B.; Rosa, J.R.B.F.; Gazaffi, R.; Marçal, J.F.; Carbonell, S.A.M.; Chiorato, A.F.; Zucchi, M.I.; Garcia, A.A.F.; Benchimol-Reis, L.L. Developing a common bean core collection suitable for association mapping studies. Genet. Mol. Biol. 2015, 38, 67–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, V.D.; Pastor-Corrales, M.A.; Chirwa, R.M.; Buruchara, R.A. Andean beans (Phaseolus vulgaris L.) with resistance to the angular leaf spot pathogen (Phaeoisariopsis griseola) in southern and eastern Africa. Euphytica 2004, 136, 201–210. [Google Scholar] [CrossRef]
- CIMMYT Laboratory Protocols: CIMMYT Applied Molecular Genetics Laboratory Protocols, 3rd ed.; CIMMYT: Mexico City, Mexico, 2005.
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Schoonhove, A.; Pastor-Corrales, M. Sistema Estándar para la Evaluación de Germoplasma de Frijol; CIAT: Cali, Colombia, 1987; ISBN 84-89206-69-4. [Google Scholar]
- CIAT Condiciones de campo para realizar las evaluaciones del germoplasma de frijol; CIAT: Cali, Colombia, 1976.
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Kalinowski, S.T. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- El Mousadik, A.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef]
- Vigouroux, Y.; McMullen, M.; Hittinger, C.T.; Houchins, K.; Schulz, L.; Kresovich, S.; Matsuoka, Y.; Doebley, J. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 2002, 99, 9650–9655. [Google Scholar] [CrossRef] [Green Version]
- Dray, S.; Dufour, A.B.; Chessel, D. The ade4 package-II: Two-table and K-table methods. R News 2007, 7, 47–52. [Google Scholar]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [Green Version]
- Francis, R.M. pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R.E. Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Bonferroni, C. Teoria statistica delle classi e calcolo delle probabilità. Pubbl. R Ist. Sup. Sci. Econ. Commer. Fir. 1963, 8, 3–62. [Google Scholar]
- Mangin, B.; Siberchicot, A.; Nicolas, S.; Doligez, A.; This, P.; Cierco-Ayrolles, C. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 2012, 108, 285–291. [Google Scholar] [CrossRef] [Green Version]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, W.G.; Weir, B.S. Variances and covariances of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 1988, 33, 54–78. [Google Scholar] [CrossRef]
- Diniz, A.L.; Giordani, W.; Costa, Z.P.; Margarido, G.R.A.; Perseguini, J.M.K.C.; Benchimol-Reis, L.L.; Chiorato, A.F.; Garcia, A.A.F.; Vieira, M.L.C. Evidence for strong kinship influence on the extent of linkage disequilibrium in cultivated common beans. Genes 2019, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Pompeu, A.S. IAC-Maravilha, IAC-Una, IAC-Carioca Pyatã, IAC-Carioca Aruã, IAC-Carioca Akytã e IAC-Bico de Ouro: Novos cultivares de feijoeiro. Bragantia 1997, 56, 79–85. [Google Scholar] [CrossRef]
- Valdisser, P.A.M.R.; Pereira, W.J.; Almeida Filho, J.E.; Müller, B.S.F.; Coelho, G.R.C.; de Menezes, I.P.P.; Vianna, J.P.G.; Zucchi, M.I.; Lanna, A.C.; Coelho, A.S.G.; et al. In-depth genome characterization of a Brazilian common bean core collection using DArTseq high-density SNP genotyping. BMC Genom. 2017, 18. [Google Scholar] [CrossRef] [Green Version]
- Navabi, A.; Balasubramanian, P.; Pauls, K.P.; Bett, K.; Hou, A. Genetic diversity of the Canadian dry bean varieties released since 1930: A pedigree analysis. Crop Sci. 2014, 54, 993–1003. [Google Scholar] [CrossRef]
- Sicard, D.; Nanni, L.; Porfiri, O.; Bulfon, D.; Papa, R. Genetic diversity of Phaseolus vulgaris L. and P. coccineus L. landraces in central Italy. Plant Breed. 2005, 124, 464–472. [Google Scholar] [CrossRef]
- Lioi, L. Variation of the storage protein phaseolin in common bean (Phaseolus vulgaris L.) from the Mediterranean area. Euphytica 1989, 44, 151–155. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Taranto, G.; Losavio, F.P.; Pignone, D. Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genet. Resour. Crop Evol. 2006, 53, 313–322. [Google Scholar] [CrossRef]
- Logozzo, G.; Donnoli, R.; Macaluso, L.; Papa, R.; Knüpffer, H.; Zeuli, P.S. Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (Phaseolus vulgaris L.) germplasm and strategies to establish a core collection. Genet. Resour. Crop Evol. 2007, 54, 1763–1779. [Google Scholar] [CrossRef]
- Cichy, K.A.; Porch, T.G.; Beaver, J.S.; Cregan, P.; Fourie, D.; Glahn, R.P.; Grusak, M.A.; Kamfwa, K.; Katuuramu, D.N.; McClean, P.; et al. A Phaseolus vulgaris diversity panel for andean bean improvement. Crop Sci. 2015, 55, 2149–2160. [Google Scholar] [CrossRef] [Green Version]
- Mamidi, S.; Rossi, M.; Moghaddam, S.M.; Annam, D.; Lee, R.; Papa, R.; McClean, P.E. Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 2013, 110, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Vega, E.; Campa, A.; De la Rosa, L.; Giraldez, R.; Ferreira, J.J. Genetic diversity in a core collection established from the main bean genebank in Spain. Crop Sci. 2009, 49, 1377–1386. [Google Scholar] [CrossRef]
- Carbonell, S.A.M.; Chiorato, A.F.; Ito, M.F.; Perina, E.F.; Gonçalves, J.G.R.; De Souza, P.S.; Gallo, P.B.; Ticelli, M.; Colombo, C.A.; De Azevedo Filho, J.A. IAC-Alvorada and IAC-Diplomata: New common bean cultivars. Crop Breed. Appl. Biotechnol. 2008, 8, 163–166. [Google Scholar] [CrossRef]
- Abreu, Â.F.B.; Ramalho, M.A.P.; Carneiro, J.E.S.; Gonçalves, F.M.A.; Santos, J.B.; Peloso, M.J.; Faria, L.C.; Carneiro, G.E.S.; Pereira Filho, I.A. ‘BRSMG Talismã’: common bean cultivar with Carioca grain type. Crop Breed. Appl. Biotechnol. 2004, 4, 372–374. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, S.A.M.; Chiorato, A.F.; Bolonhezi, D.; de Barros, V.L.N.P.; Borges, W.L.B.; Ticelli, M.; Gallo, P.B.; Finoto, E.L.; Santos, N.C.B. dos “IAC Milênio” - Common bean cultivar with high grain quality. Crop Breed. Appl. Biotechnol. 2014, 14, 273–276. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, S.A.M.; Chiorato, A.F.; Carvalho, C.R.L.; Benchimol, L.L.; Beraldo, A.L.A.; Gonçalves, J.G.R.; Ticelli, M.; De Souza, P.S.; Gallo, P.B. IAC-Galante and IAC-Centauro: Special common bean types. Crop Breed. Appl. Biotechnol. 2008, 8, 177–180. [Google Scholar] [CrossRef]
- Nay, M.M.; Souza, T.L.P.O.; Raatz, B.; Mukankusi, C.M.; Pastor-Corrales, M.A.; Abreu, A.F.B.; Melo, L.C. A review of angular leaf spot resistance in common bean. Crop Sci. 2019, 59, 1376–1391. [Google Scholar] [CrossRef] [Green Version]
- De Almeida, C.P.; Arruda, N.; de Paulino, J.F.C.; de Freitas, G.M.; Bonfante, G.F.J.; Bajay, M.M.; de Deus, B.C.; Patrício, F.R.A.; Carbonell, S.A.M.; Chiorato, A.F.; et al. Genetic diversity of Pseudocercospora griseola resistance loci in common beans. Trop. Plant Pathol. 2020, 1–10. [Google Scholar] [CrossRef]
- Bassi, D.; Briñez, B.; Rosa, J.S.; Oblessuc, P.R.; de Almeida, C.P.; Nucci, S.M.; da Silva, L.C.D.; Chiorato, A.F.; Vianello, R.P.; Camargo, L.E.A.; et al. Linkage and mapping of quantitative trait loci associated with angular leaf spot and powdery mildew resistance in common beans. Genet. Mol. Biol. 2017, 40, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves-Vidigal, M.C.; Cruz, A.S.; Garcia, A.; Kami, J.; Filho, P.S.V.; Sousa, L.L.; McClean, P.; Gepts, P.; Pastor-Corrales, M.A. Linkage mapping of the Phg-1 and Co-14 genes for resistance to angular leaf spot and anthracnose in the common bean cultivar AND 277. Theor. Appl. Genet. 2011, 122, 893–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanglard, D.A.; Mafra, V.S.; Ribeiro, C.A.G.; Da Silva, L.C.; De Barros, E.G.; Moreira, M.A.; De, J.E.; Carneiro, S.; Mota, A.P.S.; Vianello, R.P.; et al. Rudá x AND 277 RILs: A potential new core mapping population for common bean. Annu. Rep. Bean Improv. Coop. 2013, 56, 23–24. [Google Scholar]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 1–11. [Google Scholar]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Baroni, R.M.; Garcia, A.A.F.; Chioratto, A.F.; Carbonell, S.A.M.; Camargo, L.E.A.; Benchimol-Reis, L.L. Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet. 2012, 13. [Google Scholar] [CrossRef] [Green Version]
- Oblessuc, P.R.; Perseguini, J.C.M.K.; Baroni, R.M.; Chiorato, A.F.; Carbonell, S.A.M.; Mondego, J.M.C.; Vidal, R.O.; Camargo, L.E.A.; Benchimol-Reis, L.L. Increasing the density of markers around a major QTL controlling resistance to angular leaf spot in common bean. Theor. Appl. Genet. 2013, 126, 2451–2465. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, J.; Wang, L.; Wang, S. Genome-wide association study identifies NBS-LRR-encoding genes related with anthracnose and common bacterial blight in the common bean. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, P.C.B.; Brondani, C.; Menezes, I.P.P.; Valdisser, P.A.M.R.; Borba, T.C.O.; Del Peloso, M.J.; Vianello, R.P. Discrimination of common bean cultivars using multiplexed microsatellite markers. Genet. Mol. Res. 2014, 13, 1964–1978. [Google Scholar] [CrossRef]
- Bulisani, E.A. Feijão Carioca-uma História de Sucesso, 1st ed.; InfoBibos: São Paulo, Brazil, 2008. [Google Scholar]
- Allendorf, F.W. Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol. 1986, 5, 181–190. [Google Scholar] [CrossRef]
- Allendorf, F.; Luikart, G.; Aitken, S. Conservation and the Genetics of Population, 2nd ed.; Wiley-Blackwell: Malden, MA, USA, 2013. [Google Scholar]
- Vilas, A.; Pérez-Figueroa, A.; Quesada, H.; Caballero, A. Allelic diversity for neutral markers retains a higher adaptive potential for quantitative traits than expected heterozygosity. Mol. Ecol. 2015, 24, 4419–4432. [Google Scholar] [CrossRef]
- Repinski, S.L.; Kwak, M.; Gepts, P. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 2012, 124, 1539–1547. [Google Scholar] [CrossRef]
- Koinange, E.M.K.; Singh, S.P.; Gepts, P. Genetic control of the domestication syndrome in common bean. Crop Sci. 1996, 36, 1037–1045. [Google Scholar] [CrossRef] [Green Version]
- Kamfwa, K.; Cichy, K.A.; Kelly, J.D. Genome-Wide Association Study of Agronomic Traits in Common Bean. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Nascimento, M.; Nascimento, A.C.C.; Silva, F.F.E.; Barili, L.D.; Do Vale, N.M.; Carneiro, J.E.; Cruz, C.D.; Carneiro, P.C.S.; Serão, N.V.L. Quantile regression for genome-wide association study of flowering time-related traits in common bean. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [Green Version]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef]






| Group | No. | Npa | I | Rs | Ho | He | uHe | PIC |
|---|---|---|---|---|---|---|---|---|
| Bean diversity panel | 185 | - | 0.317 | - | 0.018 | 0.181 | 0.182 | 0.160 |
| Special | 25 | 89 | 0.573 | 1.990 | 0.019 | 0.403 | 0.404 | 0.309 |
| Black | 29 | 0 | 0.257 | 1.890 | 0.018 | 0.152 | 0.151 | 0.128 |
| Carioca | 131 | 6 | 0.199 | 1.650 | 0.018 | 0.115 | 0.114 | 0.099 |
| Modern Carioca | 53 | 0 | 0.198 | 1.970 | 0.017 | 0.117 | 0.118 | 0.100 |
| Old Carioca | 36 | 0 | 0.163 | 1.930 | 0.023 | 0.095 | 0.094 | 0.081 |
| Carioca lines | 42 | 2 | 0.200 | 1.980 | 0.015 | 0.118 | 0.117 | 0.101 |
| Andean | 10 | 422 | 0.152 | 1.360 | 0.002 | 0.099 | 0.100 | 0.080 |
| Mesoamerican | 175 | 1839 | 0.211 | 1.520 | 0.019 | 0.121 | 0.122 | 0.105 |
| Variance Source | df | SS | Variance | |
|---|---|---|---|---|
| Component | (%) | |||
| Between commercial classes | 2 | 12,889.0 | 71.3 | 23.86 * |
| Between cultivars | 182 | 78,172.9 | 201.9 | 67.52 * |
| Within cultivars | 185 | 4761.0 | 25.7 | 8.60 * |
| Total | 369 | 95,822.9 | 299.00 | 100 |
| Trait | Cultivar | Repetition | Residual | CV | h2 | S-P |
|---|---|---|---|---|---|---|
| Flowering time | 14.89 *** | 6.06 ns | 3.44 | 4.75% | 77.97% | 0.07 ns |
| Pod maturation | 25.69 *** | 23.79 *** | 4.45 | 2.78% | 83.15% | 0.51 ns |
| Growth habit | 0.58 *** | 1.28 *** | 0.93 | 11.04% | 83.09% | 0.0001 *** |
| df | 140 | 3 | 420 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, C.P.d.; Paulino, J.F.d.C.; Morais Carbonell, S.A.; Chiorato, A.F.; Song, Q.; Di Vittori, V.; Rodriguez, M.; Papa, R.; Benchimol-Reis, L.L. Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding. Genes 2020, 11, 1298. https://doi.org/10.3390/genes11111298
Almeida CPd, Paulino JFdC, Morais Carbonell SA, Chiorato AF, Song Q, Di Vittori V, Rodriguez M, Papa R, Benchimol-Reis LL. Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding. Genes. 2020; 11(11):1298. https://doi.org/10.3390/genes11111298
Chicago/Turabian StyleAlmeida, Caléo Panhoca de, Jean Fausto de Carvalho Paulino, Sérgio Augusto Morais Carbonell, Alisson Fernando Chiorato, Qijian Song, Valerio Di Vittori, Monica Rodriguez, Roberto Papa, and Luciana Lasry Benchimol-Reis. 2020. "Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding" Genes 11, no. 11: 1298. https://doi.org/10.3390/genes11111298
APA StyleAlmeida, C. P. d., Paulino, J. F. d. C., Morais Carbonell, S. A., Chiorato, A. F., Song, Q., Di Vittori, V., Rodriguez, M., Papa, R., & Benchimol-Reis, L. L. (2020). Genetic Diversity, Population Structure, and Andean Introgression in Brazilian Common Bean Cultivars after Half a Century of Genetic Breeding. Genes, 11(11), 1298. https://doi.org/10.3390/genes11111298








