All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers
Abstract
:1. Introduction
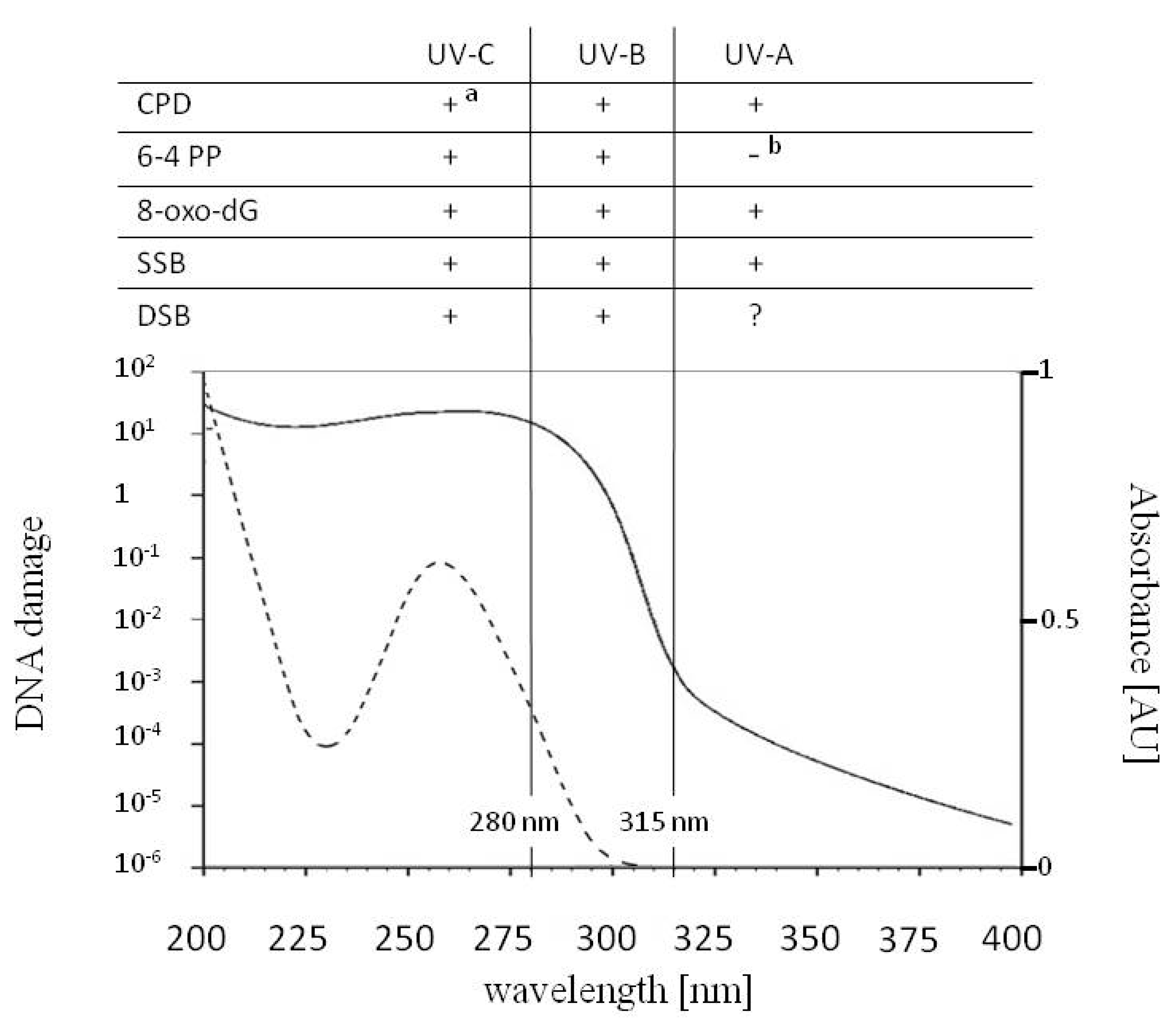
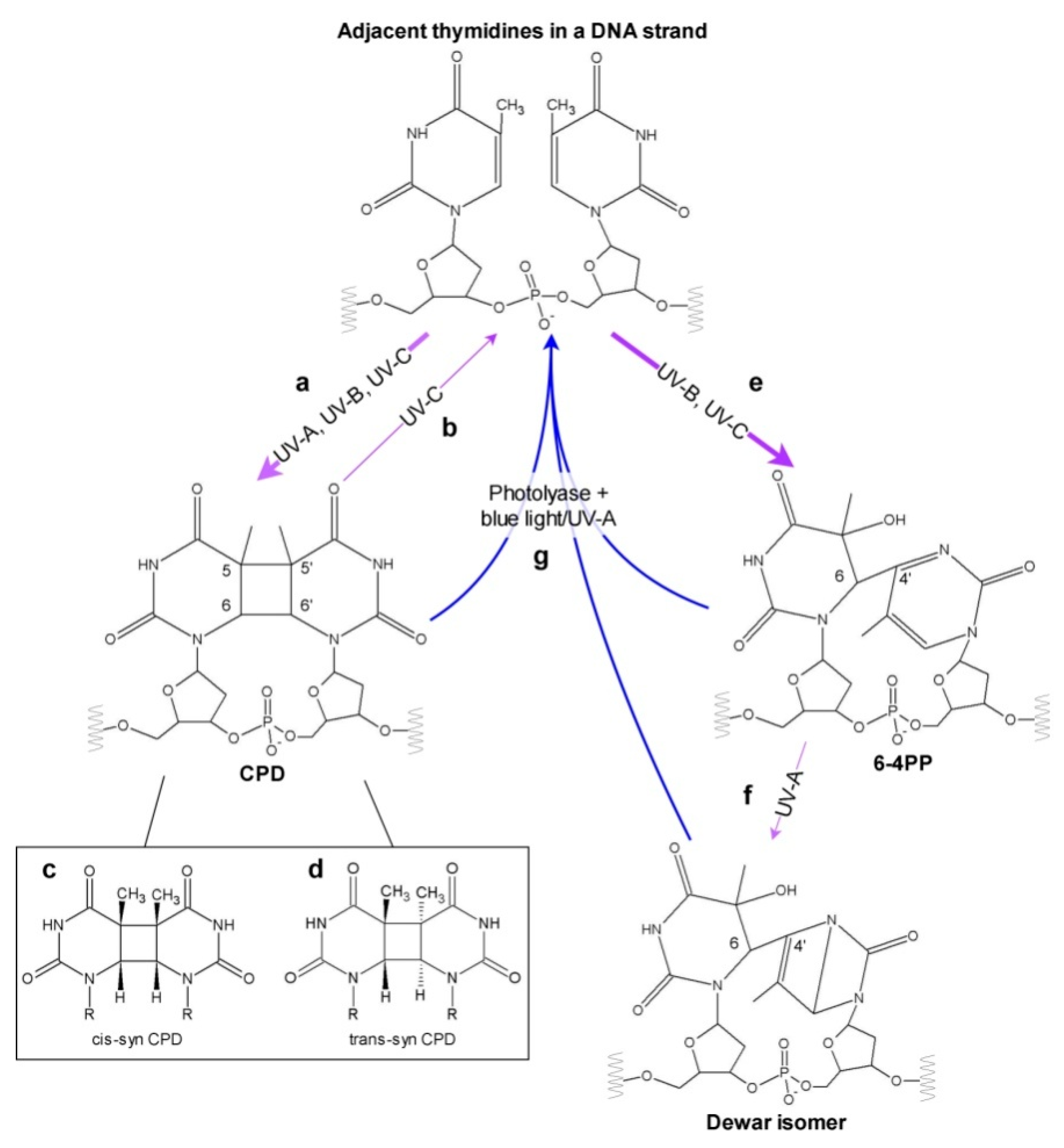
2. UV Impact on DNA
3. Photoreactivation
4. Perspectives
4.1. DNA Packaging, Metabolism and Interaction between Light and Dark Repair
4.2. Repair of UV-Induced Lesions in Chloroplasts
4.3. The Role of Phosphorylation in Photorepair
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ATM | Ataxia telangiectasia-mutated |
| ATR | Ataxia telangiectasia-mutated and Rad3-related |
| CHR8 | chromatin remodeling 8 |
| CPD | cyclobutane pyrimidine dimer |
| CRY | cryptochrome |
| CRY-DASH | Cryptochrome-Drosophila, Arabidopsis, Synechocystis, Human proteins |
| DMRL | 6,7-dimethyl-8-ribityl-lumazin |
| DSB | double strand breaks |
| FAD | flavin adenine dinucleotide |
| MTHF | 5,10-methenyltetrahydrofolate |
| NER | nucleotide excision repair |
| PHR | photolyase homology region |
| ROS | reactive oxygen species |
| SSB | single strand break |
| UV | ultraviolet |
| 6-4 PP | (6-4) pyrimidine–pyrimidone photoproduct |
| 8-HDF | 8-hydroxydeazaflavin |
References
- Fu, Q. Radiation (SOLAR). In Encyclopedia of Atmospheric Sciences; Holton, J.R., Ed.; Academic Press: Oxford, UK, 2003; pp. 1859–1863. [Google Scholar]
- CIE. The spectroradiometric measurement of light sources. In International Commission on Illumination; CIE: Vienna, Austria, 1984; Volume 63. [Google Scholar]
- CIE. The measurement of absolute luminous intensity distributions. In International Commission on Illumination; CIE: Vienna, Austria, 1987; Volume 70. [Google Scholar]
- CIE. Standardization of the Terms UV-A1, UV-A2 and UV-B. In International Commission on Illumination; CIE: Vienna, Austria, 1999; Volume 134. [Google Scholar]
- Douki, T. Pyrimidine (6-4) Pyrimidone Photoproducts in UVA-Irradiated DNA: Photosensitization or Photoisomerization? ChemPhotoChem 2020, 4, 294–299. [Google Scholar] [CrossRef]
- Molina, L.T.; Molina, M.J. Absolute absorption cross sections of ozone in the 185- to 350-nm wavelength range. J. Geophys. Res. 1986, 91. [Google Scholar] [CrossRef]
- Moan, J. Visible Light and UV Radiation. In Radiation at Home, Outdoors and in the Workplace; Scandinavian Science Publisher: Oslo, Norway, 2002; Chapter 7 & 31. [Google Scholar]
- Neves-Petersen, T.M.; Prakash, G.; Petersen, S. UV Light Effects on Proteins: From Photochemistry to Nanomedicine. In Molecular Photochemistry - Various Aspects; Saha, S., Ed.; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef] [Green Version]
- Casati, P.; Walbot, V. Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant. Physiol. 2004, 136, 3319–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockell, C.S. Biological effects of high ultraviolet radiation on early earth—A theoretical evaluation. J. Theor. Biol. 1998, 193, 717–729. [Google Scholar] [CrossRef]
- Fischer, J.H. Specific detection of nucleotides, creatine phosphate, and their derivatives from tissue samples in a simple, isocratic, recycling, low-volume system. LC-GC Int.—Mag. Sep. Sci. 1995, 8, 254–264. [Google Scholar]
- Basu, S. Ultraviolet absorption studies on DNA. Biopolymers 1967, 5, 876–878. [Google Scholar] [CrossRef] [PubMed]
- Yagura, T.; Makita, K.; Yamamoto, H.; Menck, C.F.M.; Schuch, A.P. Biological sensors for solar ultraviolet radiation. Sensors 2011, 11, 4277–4294. [Google Scholar] [CrossRef] [Green Version]
- Beck, S.E.; Rodriguez, R.A.; Linden, K.G.; Hargy, T.M.; Larason, T.C.; Wright, H.B. Wavelength dependent UV inactivation and DNA damage of adenovirus as measured by cell culture infectivity and long range quantitative PCR. Environ. Sci. Technol. 2014, 48, 591–598. [Google Scholar] [CrossRef]
- Schwalb, N.K.; Temps, F. Base Sequence and Higher-Order Structure Induce the Complex Excited-State Dynamics in DNA. Science 2008, 322, 243–245. [Google Scholar] [CrossRef]
- Felsenfeld, G.; Hirschman, S.Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J. Mol. Biol. 1965, 13, 407–427. [Google Scholar] [CrossRef]
- Markovitsi, D. UV-induced DNA Damage: The Role of Electronic Excited States. Photochem. Photobiol. 2016, 92, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setlow, R.B. The wavelengths in sunlight effective in producing skin cancer: A theoretical analysis. Proc. Natl. Acad. Sci. USA 1974, 71, 3363–3366. [Google Scholar] [CrossRef] [Green Version]
- Kielbassa, C.; Roza, L.; Epe, B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis 1997, 18, 811–816. [Google Scholar] [CrossRef] [Green Version]
- Dany, A.L.; Douki, T.; Triantaphylides, C.; Cadet, J. Repair of the main UV-induced thymine dimeric lesions within Arabidopsis thaliana DNA: Evidence for the major involvement of photoreactivation pathways. J. Photochem. Photobiol. B Biol. 2001, 65, 127–135. [Google Scholar] [CrossRef]
- Douki, T.; Cadet, J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry 2001, 40, 2495–2501. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.L.; Jen, J.; Cleaver, J.E. Sequence specificity of cyclobutane pyrimidine dimers in DNA treated with solar (ultraviolet B) radiation. Nucleic Acids Res. 1992, 20, 225–229. [Google Scholar] [CrossRef] [Green Version]
- Rochette, P.J.; Therrien, J.P.; Drouin, R.; Perdiz, D.; Bastien, N.; Drobetsky, E.A.; Sage, E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003, 31, 2786–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sage, E. Distribution and repair of photolesions in DNA: Genetic consequences and the role of sequence context. Photochem. Photobiol. 1993, 57, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Clivio, P.; Guillaume, D. Far UV irradiation of a bis-thymine PNA dimer: Conformational implications. Tetrahedron Lett. 1998, 39, 6881–6884. [Google Scholar] [CrossRef]
- Douki, T. The variety of UV-induced pyrimidine dimeric photoproducts in DNA as shown by chromatographic quantification methods. Photochem. Photobiol. Sci. 2013, 12, 1286–1302. [Google Scholar] [CrossRef]
- Law, Y.K.; Forties, R.A.; Liu, X.; Poirier, M.G.; Kohler, B. Sequence-dependent thymine dimer formation and photoreversal rates in double-stranded DNA. Photochem. Photobiol. Sci. 2013, 12, 1431–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeifer, G.P.; Drouin, R.; Riggs, A.D.; Holmquist, G.P. In vivo mapping of a DNA adduct at nucleotide resolution: Detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1991, 88, 1374–1378. [Google Scholar] [CrossRef] [Green Version]
- Sancar, A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003, 103, 2203–2237. [Google Scholar] [CrossRef] [PubMed]
- Douki, T.; Sage, E. Dewar valence isomers, the third type of environmentally relevant DNA photoproducts induced by solar radiation. Photochem. Photobiol. Sci. 2016, 15, 24–30. [Google Scholar] [CrossRef]
- Haiser, K.; Fingerhut, B.P.; Heil, K.; Glas, A.; Herzog, T.T.; Pilles, B.M.; Schreier, W.J.; Zinth, W.; Devivie-Riedle, R.; Carell, T. Mechanism of UV-induced formation of Dewar lesions in DNA. Angew. Chem.—Int. Ed. 2012, 51, 408–411. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Murakami, M.; Nakajima, N.; Kondo, N.; Nikaido, O. The photorepair and photoisomerization of DNA lesions in etiolated cucumber cotyledons after irradiation by UV-B depends on wavelength. Plant Cell Physiol. 1998, 39, 745–750. [Google Scholar] [CrossRef]
- Cuquerella, C.; Lhiaubet-Vallet, V.; Bosca, F.; Miranda, M.A. Photosensitised pyrimidine dimerisation in DNA. Chem. Sci. 2011, 2, 1219–1232. [Google Scholar] [CrossRef]
- Vendrell-Criado, V.; Rodríguez-Muñiz, G.M.; Lhiaubet-Vallet, V.; Cuquerella, M.C.; Miranda, M.A. The (6–4) Dimeric Lesion as a DNA Photosensitizer. ChemPhysChem 2016, 17, 1979–1982. [Google Scholar] [CrossRef] [Green Version]
- Premi, S.; Wallisch, S.; Mano, C.M.; Weiner, A.B.; Bacchiocchi, A.; Wakamatsu, K.; Bechara, E.J.H.; Halaban, R.; Douki, T.; Brash, D.E. Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 2015, 346, 842–848. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Gasparutto, D.; Ravanat, J.L. Oxidative damage to DNA: Formation, measurement and biochemical features. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2003, 531, 5–23. [Google Scholar] [CrossRef]
- Balestrazzi, A.; Locato, V.; Bottone, M.G.; De Gara, L.; Biggiogera, M.; Pellicciari, C.; Botti, S.; Di Ges, D.; Don, M.; Carbonera, D. Response to UV-C radiation in topo I-deficient carrot cells with low ascorbate levels. J. Exp. Bot. 2010, 61, 575–585. [Google Scholar] [CrossRef]
- Watanabe, K.; Yamada, N.; Takeuchi, Y. Oxidative DNA damage in cucumber cotyledons irradiated with ultraviolet light. J. Plant. Res. 2006, 119, 239–246. [Google Scholar] [CrossRef]
- Peak, J.G.; Peak, M.J. Ultraviolet light induces double-strand breaks in DNA of cultured human P3 cells as measured by neutral filter elution. Photochem. Photobiol. 1990, 52, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.L.; Dunn, J.; Rees, A.; Rünger, T.M. No formation of DNA double-strand breaks and no activation of recombination repair with UVA. J. Investig. Dermatol. 2011, 131, 1139–1148. [Google Scholar] [CrossRef] [Green Version]
- Svobodová, A.R.; Galandáková, A.; Šianská, J.; Doležal, D.; Lichnovská, R.; Ulrichová, J.; Vostálová, J. DNA damage after acute exposure of mice skin to physiological doses of UVB and UVA light. Arch. Dermatol. Res. 2012, 304, 407–412. [Google Scholar] [CrossRef]
- Greinert, R.; Volkmer, B.; Henning, S.; Breitbart, E.W.; Greulich, K.O.; Cardoso, M.C.; Rapp, A. UVA-induced DNA double-strand breaks result from the repair of clustered oxidative DNA damages. Nucleic Acids Res. 2012, 40, 10263–10273. [Google Scholar] [CrossRef] [Green Version]
- Besaratinia, A.; Yoon, J.; Schroeder, C.; Bradforth, S.E.; Cockburn, M.; Pfeifer, G.P. Wavelength dependence of ultraviolet radiation-induced DNA damage as determined by laser irradiation suggests that cyclobutane pyrimidine dimers are the principal DNA lesions produced by terrestrial sunlight. FASEB J. 2011, 25, 3079–3091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douki, T.; Reynaud-Angelin, A.; Cadet, J.; Sage, E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry 2003, 42, 9221–9226. [Google Scholar] [CrossRef] [PubMed]
- Douki, T.; Perdiz, D.; Gróf, P.; Kuluncsics, Z.; Moustacchi, E.; Cadet, J.; Sage, E. Oxidation of guanine in cellular DNA by solar UV radiation: Biological role. Photochem. Photobiol. 1999, 70, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Kucera, B.; Leubner-Metzger, G.; Wellmann, E. Distinct Ultraviolet-Signaling Pathways in Bean Leaves. DNA Damage Is Associated with β-1,3-Glucanase Gene Induction, but Not with Flavonoid Formation. Plant. Physiol. 2003, 133, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Ioki, M.; Takahashi, S.; Nakajima, N.; Saji, H.; Fujikura, K.; Tamaoki, M.; Aono, M.; Kanna, M.; Ogawa, D.; Watanabe, M.; et al. Wavelength dependency of the light-driven transcriptional activation of the cucumber CPD photolyase gene. Phyton Ann. Rei Bot. 2005, 45, 177–184. [Google Scholar]
- Biever, J.J.; Brinkman, D.; Gardner, G. UV-B inhibition of hypocotyl growth in etiolated Arabidopsis thaliana seedlings is a consequence of cell cycle arrest initiated by photodimer accumulation. J. Exp. Bot. 2014, 65, 2949–2961. [Google Scholar] [CrossRef] [Green Version]
- Douki, T. Effect of denaturation on the photochemistry of pyrimidine bases in isolated DNA. J. Photochem. Photobiol. B Biol. 2006, 82, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Murakami, M.; Nakajima, N.; Kondo, N.; Nikaido, O. Induction and repair of damage to DNA in cucumber cotyledons irradiated with UV-B. Plant. Cell Physiol. 1996, 37, 181–187. [Google Scholar] [CrossRef]
- Li, S.; Paulsson, M.; Björn, L.O. Temperature-dependent formation and photorepair of DNA damage induced by UV-B radiation in suspension-cultured tobacco cells. J. Photochem. Photobiol. B Biol. 2002, 66, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wang, Y.; Björn, L.O. Effects of temperature on UV-B-induced DNA damage and photorepair in Arabidopsis thaliana. J. Environ. Sci. 2004, 16, 173–176. [Google Scholar]
- Park, H.J.; Zhang, K.; Ren, Y.; Nadji, S.; Sinha, N.; Taylor, J.S.; Kang, C.H. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. USA 2002, 99, 15965–15970. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, H.; Mizutani, R. Structural biology of DNA (6-4) photoproducts formed by ultraviolet radiation and interactions with their binding proteins. Int. J. Mol. Sci. 2014, 15, 20321–20338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, A.; Tachiwana, H.; Kagawa, W.; Horikoshi, N.; Matsumoto, S.; Hasegawa, M.; Matsumoto, N.; Toga, T.; Yamamoto, J.; Hanaoka, F.; et al. Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome. Sci. Rep. 2015, 5, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horikoshi, N.; Tachiwana, H.; Kagawa, W.; Osakabe, A.; Matsumoto, S.; Iwai, S.; Sugasawa, K.; Kurumizaka, H. Crystal structure of the nucleosome containing ultraviolet light-induced cyclobutane pyrimidine dimer. Biochem. Biophys. Res. Commun. 2016, 471, 117–122. [Google Scholar] [CrossRef]
- Chan, G.L.; Doetsch, P.W.; Haseltine, W.A. Cyclobutane Pyrimidine Dimers and (6-4) Photoproducts Block Polymerization by DNA Polymerase I. Biochemistry 1985, 24, 5723–5728. [Google Scholar] [CrossRef] [PubMed]
- Mei Kwei, J.S.; Kuraoka, I.; Horibata, K.; Ubukata, M.; Kobatake, E.; Iwai, S.; Handa, H.; Tanaka, K. Blockage of RNA polymerase II at a cyclobutane pyrimidine dimer and 6-4 photoproduct. Biochem. Biophys. Res. Commun. 2004, 320, 1133–1138. [Google Scholar] [CrossRef]
- You, Y.H.; Lee, D.H.; Yoon, J.H.; Nakajima, S.; Yasui, A.; Pfeifer, G.P. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J. Biol. Chem. 2001, 276, 44688–44694. [Google Scholar] [CrossRef] [Green Version]
- Kelner, A. Effect of Visible Light on the Recovery of Streptomyces Griseus Conidia from Ultra-violet Irradiation Injury. Proc. Natl. Acad. Sci. USA 1949, 35, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Rupert, C.S.; Goodgal, S.H.; Herriot, R.M. Photoreactivation in vitro of ultraviolet-inactivated Hemophilus influenzae transforming factor. J. Gen. Physiol. 1958, 41, 451–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas-Lledó, J.I.; Lynch, M. Evolution of mutation rates: Phylogenomic analysis of the photolyase/cryptochrome family. Mol. Biol. Evol. 2009, 26, 1143–1153. [Google Scholar] [CrossRef] [Green Version]
- van Noort, S.J.T.; van der Werf, K.O.; Eker, A.P.M.; Wyman, C.; De Grooth, B.G.; Van Hulst, N.F.; Greve, J. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys. J. 1998, 74, 2840–2849. [Google Scholar] [CrossRef] [Green Version]
- Husain, I.; Sancar, A. Binding of E. coli DNA photolyase to a defined substrate containing a single T mean value of T dimer. Nucleic Acids Res. 1987, 15, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Sancar, G.B.; Smith, F.W.; Sancar, A. Binding of Escherichia coli DNA Photolyase to UV-Irradiated DNA. Biochemistry 1985, 24, 1849–1855. [Google Scholar] [CrossRef]
- Vande Berg, B.J.; Sancar, G.B. Evidence for dinucleotide flipping by DNA photolyase. J. Biol. Chem. 1998, 273, 20276–20284. [Google Scholar] [CrossRef] [Green Version]
- Van Noort, J.; Orsini, F.; Eker, A.; Wyman, C.; de Grooth, B.; Greve, J. DNA bending by photolyase in specific and non-specific complexes studied by atomic force microscopy. Nucleic Acids Res. 1999, 27, 3875–3880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knips, A.; Zacharias, M. Both DNA global deformation and repair enzyme contacts mediate flipping of thymine dimer damage. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marizcurrena, J.J.; Acosta, S.; Canclini, L.; Hernández, P.; Vallés, D.; Lamparter, T.; Castro-Sowinski, S. A natural occurring bifunctional CPD/(6-4)-photolyase from the Antarctic bacterium Sphingomonas sp. UV9. Appl. Microbiol. Biotechnol. 2020, 104, 7037–7050. [Google Scholar] [CrossRef]
- Fingerhut, B.P.; Heil, K.; Kaya, E.; Oesterling, S.; De Vivie-Riedle, R.; Carell, T. Mechanism of UV-induced Dewar lesion repair catalysed by DNA (6-4) photolyase. Chem. Sci. 2012, 3, 1794–1797. [Google Scholar] [CrossRef]
- Yamada, D.; Dokainish, H.M.; Iwata, T.; Yamamoto, J.; Ishikawa, T.; Todo, T.; Iwai, S.; Getzoff, E.D.; Kitao, A.; Kandori, H. Functional Conversion of CPD and (6-4) Photolyases by Mutation. Biochemistry 2016, 55, 4173–4183. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, N. Phylogenetic and Functional Classification of the Photolyase/Cryptochrome Family. Photochem. Photobiol. 2017, 93, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Baxter, R.; Smith, B.S.; Partch, C.L.; Colbert, C.L.; Deisenhofer, J. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17701–17706. [Google Scholar] [CrossRef] [Green Version]
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokorny, R.; Klar, T.; Hennecke, U.; Carell, T.; Batschauer, A.; Essen, L.O. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc. Natl. Acad. Sci. USA 2008, 105, 21023–21027. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, L.; Zhong, D. Dynamics and mechanisms of DNA repair by photolyase. Phys. Chem. Chem. Phys. 2015, 17, 11933–11949. [Google Scholar] [CrossRef] [Green Version]
- Kiontke, S.; Gnau, P.; Haselsberger, R.; Batschauer, A.; Essen, L.O. Structural and evolutionary aspects of antenna chromophore usage by class II photolyases. J. Biol. Chem. 2014, 289, 19659–19669. [Google Scholar] [CrossRef] [Green Version]
- Oberpichler, I.; Pierik, A.J.; Wesslowski, J.; Pokorny, R.; Rosen, R.; Vugman, M.; Zhang, F.; Neubauer, O.; Ron, E.Z.; Batschauer, A.; et al. A photolyase-like protein from agrobacterium tumefaciens with an iron-sulfur cluster. PLoS ONE 2011, 6, 2–11. [Google Scholar] [CrossRef]
- Kleiner, O.; Butenandt, J.; Carell, T.; Batschauer, A. Class II DNA photolyase from Arabidopsis thaliana contains FAD as a cofactor. Eur. J. Biochem. 1999, 264, 161–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirouchi, T.; Nakajima, S.; Najrana, T.; Tanaka, M.; Matsunaga, T.; Hidema, J.; Teranishi, M.; Fujino, T.; Kumagai, T.; Yamamoto, K. A gene for a class II DNA photolyase from Oryza sativa: Cloning of the cDNA by dilution-amplification. Mol. Genet. Genom. 2003, 269, 508–516. [Google Scholar] [CrossRef]
- Teranishi, M.; Nakamura, K.; Morioka, H.; Yamamoto, K.; Hidema, J. The native cyclobutane pyrimidine dimer photolyase of rice is phosphorylated. Plant. Physiol. 2008, 146, 1941–1951. [Google Scholar] [CrossRef] [Green Version]
- Pang, Q.; Hays, J.B. UV-B-inducible and temperature-sensitive photoreactivation of cyclobutane pyrimidine dimers in Arabidopsis thaliana. Plant. Physiol. 1991, 95, 536–543. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, S.; Sugiyama, M.; Iwai, S.; Hitomi, K.; Otoshi, E.; Kim, S.T.; Jiang, C.Z.; Todo, T.; Britt, A.B.; Yamamoto, K. Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998, 26, 638–644. [Google Scholar] [CrossRef] [Green Version]
- Hada, M.; Hino, K.; Buchholz, G.; Goss, J.; Wellmann, E.; Shin, M. Assay of DNA photolyase activity in spinach leaves in relation to cell compartmentation-evidence for lack of DNA photolyase in chloroplasts. Biosci. Biotechnol. Biochem. 2000, 64, 1288–1291. [Google Scholar] [CrossRef] [PubMed]
- Göbel, T.; Reisbacher, S.; Batschauer, A.; Pokorny, R. Flavin Adenine Dinucleotide and N5,N10-Methenyltetrahydrofolate are the in planta Cofactors of Arabidopsis thaliana Cryptochrome 3. Photochem. Photobiol. 2017, 93, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Maul, M.J.; Barends, T.R.M.; Glas, A.F.; Cryle, M.J.; Domratcheva, T.; Schneider, S.; Schlichting, I.; Carell, T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. 2008, 47, 10076–10080. [Google Scholar] [CrossRef]
- Mei, Q.; Dvornyk, V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Burney, S.; Wenzel, R.; Kottke, T.; Roussel, T.; Hoang, N.; Bouly, J.-P.; Bittl, R.; Heberle, J.; Ahmad, M. Single Amino Acid Substitution Reveals Latent Photolyase Activity in Arabidopsis cry1. Angew. Chem. 2012, 124, 9490–9494. [Google Scholar] [CrossRef]
- Xu, L.; Wen, B.; Wang, Y.; Tian, C.; Wu, M.; Zhu, G. Residues at a Single Site Differentiate Animal Cryptochromes from Cyclobutane Pyrimidine Dimer Photolyases by Affecting the Proteins’ Preferences for Reduced FAD. ChemBioChem 2017, 18, 1129–1137. [Google Scholar] [CrossRef]
- Quaite, F.E.; Takayanagi, S.; Ruffini, J.; Sutherland, J.C.; Sutherland, B.M. DNA damage levels determine cyclobutyl pyrimidine dimer repair mechanisms in alfalfa seedlings. Plant Cell 1994, 6, 1635–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, S.; Tahira, Y.; Ishibashi, T.; Mori, Y.; Mori, T.; Hashimoto, J.; Sakaguchi, K. DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res. 2004, 32, 2760–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidema, J.; Taguchi, T.; Ono, T.; Teranishi, M.; Yamamoto, K.; Kumagai, T. Increase in CPD photolyase activity functions effectively to prevent growth inhibition caused by UVB radiation. Plant. J. 2007, 50, 70–79. [Google Scholar] [CrossRef]
- Kaiser, G.; Kleiner, O.; Beisswenger, C.; Batschauer, A. Increased DNA repair in Arabidopsis plants overexpressing CPD photolyase. Planta 2009, 230, 505–515. [Google Scholar] [CrossRef]
- Willing, E.M.; Piofczyk, T.; Albert, A.; Winkler, J.B.; Schneeberger, K.; Pecinka, A. UVR2 ensures transgenerational genome stability under simulated natural UV-B in Arabidopsis thaliana. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—from models to crops. Front. Plant. Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strzalka, W.; Zglobicki, P.; Bazant, A.; Kowalska, E.; Dziga, D.; Banas, A.K. The dark side of the UV induced lesions repair. Genes 2020. under review. [Google Scholar]
- Gaillard, H.; Fitzgerald, D.J.; Smith, C.L.; Peterson, C.L.; Richmond, T.J.; Thoma, F. Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J. Biol. Chem. 2003, 278, 17655–17663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suter, B.; Livingstone-Zatchej, M.; Thoma, F. Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J. 1997, 16, 2150–2160. [Google Scholar] [CrossRef] [Green Version]
- Casati, P.; Campi, M.; Chu, F.; Suzuki, N.; Maltby, D.; Guan, S.; Burlingame, A.L.; Walbot, V. Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant. Cell 2008, 20, 827–842. [Google Scholar] [CrossRef] [Green Version]
- Graindorge, S.; Cognat, V.; Berens, P.J.; Mutterer, J.; Molinier, J. Photodamage repair pathways contribute to the accurate maintenance of the DNA methylome landscape upon UV exposure. PLoS Genet. 2019, 15, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Livingstone-Zatchej, M.; Meier, A.; Suter, B.; Thoma, F. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 1997, 25, 3795–3800. [Google Scholar] [CrossRef] [Green Version]
- Svejstrup, J.Q. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 2003, 116, 447–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citterio, E.; Van Den Boom, V.; Schnitzler, G.; Kanaar, R.; Bonte, E.; Kingston, R.E.; Hoeijmakers, J.H.J.; Vermeulen, W. ATP-Dependent Chromatin Remodeling by the Cockayne Syndrome B DNA Repair-Transcription-Coupling Factor. Mol. Cell. Biol. 2000, 20, 7643–7653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Khateeb, W.M.; Sher, A.A.; Marcus, J.M.; Schroeder, D.F. UVSSA, UBP12, and RDO2/TFIIS contribute to Arabidopsis UV tolerance. Front. Plant Sci. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Sancar, G.B.; Smith, F.W. Interactions between yeast photolyase and nucleotide excision repair proteins in Saccharomyces cerevisiae and Escherichia coli. Mol. Cell. Biol. 1989, 9, 4767–4776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, M.E.; Feldman, B.J.; Chu, G. A novel role for DNA photolyase: Binding to DNA damaged by drugs is associated with enhanced cytotoxicity in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994, 14, 8071–8077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özer, Z.; Reardon, J.T.; Hsu, D.S.; Malhotra, K.; Sancar, A. The other function of DNA photolyase: Stimulation of excision repair of chemical damage to DNA. Biochemistry 1995, 34, 15886–15889. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, T.; Curtis, M.J.; Tominey, C.M.; Duong, Y.H.; Wilcox, B.W.L.; Aggoune, D.; Hays, J.B.; Britt, A.B. A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst.) 2010, 9, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.J.; Hays, J.B. Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair (Amst.) 2011, 10, 1272–1281. [Google Scholar] [CrossRef]
- Hung, K.F.; Sidorova, J.M.; Nghiem, P.; Kawasumi, M. The 6-4 photoproduct is the trigger of UV-induced replication blockage and ATR activation. Proc. Natl. Acad. Sci. USA 2020, 117, 12806–12816. [Google Scholar] [CrossRef] [PubMed]
- Banas, A.K.; Hermanowicz, P.; Sztatelman, O.; Labuz, J.; Aggarwal, C.; Zglobicki, P.; Jagiello-Flasinska, D.; Strzalka, W. 6,4-PP Photolyase Encoded by AtUVR3 is Localized in Nuclei, Chloroplasts and Mitochondria and its Expression is Down-Regulated by Light in a Photosynthesis-Dependent Manner. Plant. Cell Physiol. 2018, 59, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Teranishi, M.; Ishida, H.; Kawasaki, J.; Takeuchi, A.; Yamaya, T.; Watanabe, M.; Makino, A.; Hidema, J. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA. Plant J. 2011, 66, 433–442. [Google Scholar] [CrossRef]
- Kleine, T.; Lockhart, P.; Batschauer, A. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant. J. 2003, 35, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Hashimoto, T.; Nikaido, O.; Shin, M. UVB-Induced DNA Damage and Its Photorepair in Nuclei and Chloroplasts of Spinacia oleracea L. Photochem. Photobiol. 1998, 68, 319–322. [Google Scholar] [CrossRef]
- Chen, J.J.; Mitchell, D.L.; Britt, A.B. A light-dependent pathway for the elimination of UV-induced pyrimidine (6-4) pyrimidinone photoproducts in Arabidopsis. Plant Cell 1994, 6, 1311–1317. [Google Scholar] [CrossRef]
- Cannon, G.C.; Hedrick, L.A.; Heinhorst, S. Repair mechanisms of UV-induced DNA damage in soybean chloroplasts. Plant Mol. Biol. 1995, 29, 1267–1277. [Google Scholar] [CrossRef]
- Draper, C.K.; Hays, J.B. Replication of chloroplast, mithochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 2000, 23, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Teranishi, M.; Nakamura, K.; Furukawa, H.; Hidema, J. Identification of a phosphorylation site in cyclobutane pyrimidine dimer photolyase of rice. Plant Physiol. Biochem. 2013, 63, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Nakajima, N.; Saji, H.; Kondo, N. Diurnal change of cucumber CPD photolyase gene (CsPHR) expression and its physiological role in growth under UV-B irradiation. Plant Cell Physiol. 2002, 43, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Waterworth, W.M.; Jiang, Q.; West, C.E.; Nikaido, M.; Bray, C.M. Characterization of Arabidopsis photolyase enzymes and analysis of their role in protection from ultraviolet-B radiation. J. Exp. Bot. 2002, 53, 1005–1015. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Sakamoto, A.; Ishigaki, Y.; Nikaido, O.; Sun, G.; Hase, Y.; Shikazono, N.; Tano, S.; Watanabe, H. An ultraviolet-B-resistant mutant with enhanced DNA repair in Arabidopsis. Plant Physiol. 2002, 129, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Hitomi, K.; Arvai, A.S.; Yamamoto, J.; Hitomi, C.; Teranishi, M.; Hirouchi, T.; Yamamoto, K.; Iwai, S.; Tainer, J.A.; Hidema, J.; et al. Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J. Biol. Chem. 2012, 287, 12060–12069. [Google Scholar] [CrossRef] [Green Version]
- Hitomi, K.; DiTacchio, L.; Arvai, A.S.; Yamamoto, J.; Kim, S.T.; Todo, T.; Tainer, J.A.; Iwai, S.; Panda, S.; Getzoff, E.D. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc. Natl. Acad. Sci. USA 2009, 106, 6962–6967. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banaś, A.K.; Zgłobicki, P.; Kowalska, E.; Bażant, A.; Dziga, D.; Strzałka, W. All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers. Genes 2020, 11, 1304. https://doi.org/10.3390/genes11111304
Banaś AK, Zgłobicki P, Kowalska E, Bażant A, Dziga D, Strzałka W. All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers. Genes. 2020; 11(11):1304. https://doi.org/10.3390/genes11111304
Chicago/Turabian StyleBanaś, Agnieszka Katarzyna, Piotr Zgłobicki, Ewa Kowalska, Aneta Bażant, Dariusz Dziga, and Wojciech Strzałka. 2020. "All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers" Genes 11, no. 11: 1304. https://doi.org/10.3390/genes11111304
APA StyleBanaś, A. K., Zgłobicki, P., Kowalska, E., Bażant, A., Dziga, D., & Strzałka, W. (2020). All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers. Genes, 11(11), 1304. https://doi.org/10.3390/genes11111304




