FLNC Expression Level Influences the Activity of TEAD-YAP/TAZ Signaling
Abstract
1. Introduction
2. Material and Methods
2.1. Generation of Flnc Knock-Out Cell Line Using CRISPR/Cas9
2.2. Cell Culture
2.3. Western Blotting
2.4. Immunofluorescence
2.5. Gene Expression Analysis
2.6. Plasmid Construction and Flnc Overexpression
2.7. Scratch Assay
2.8. Proliferation and Migration Analyses
2.9. Luciferase Assay
2.10. Statistical Analysis
3. Results
3.1. Generation and Characterization of FlncKO Cell Line
3.2. FlncKO Myoblasts Exhibit Increased Proliferation Dynamic and Reduced Migration Ability
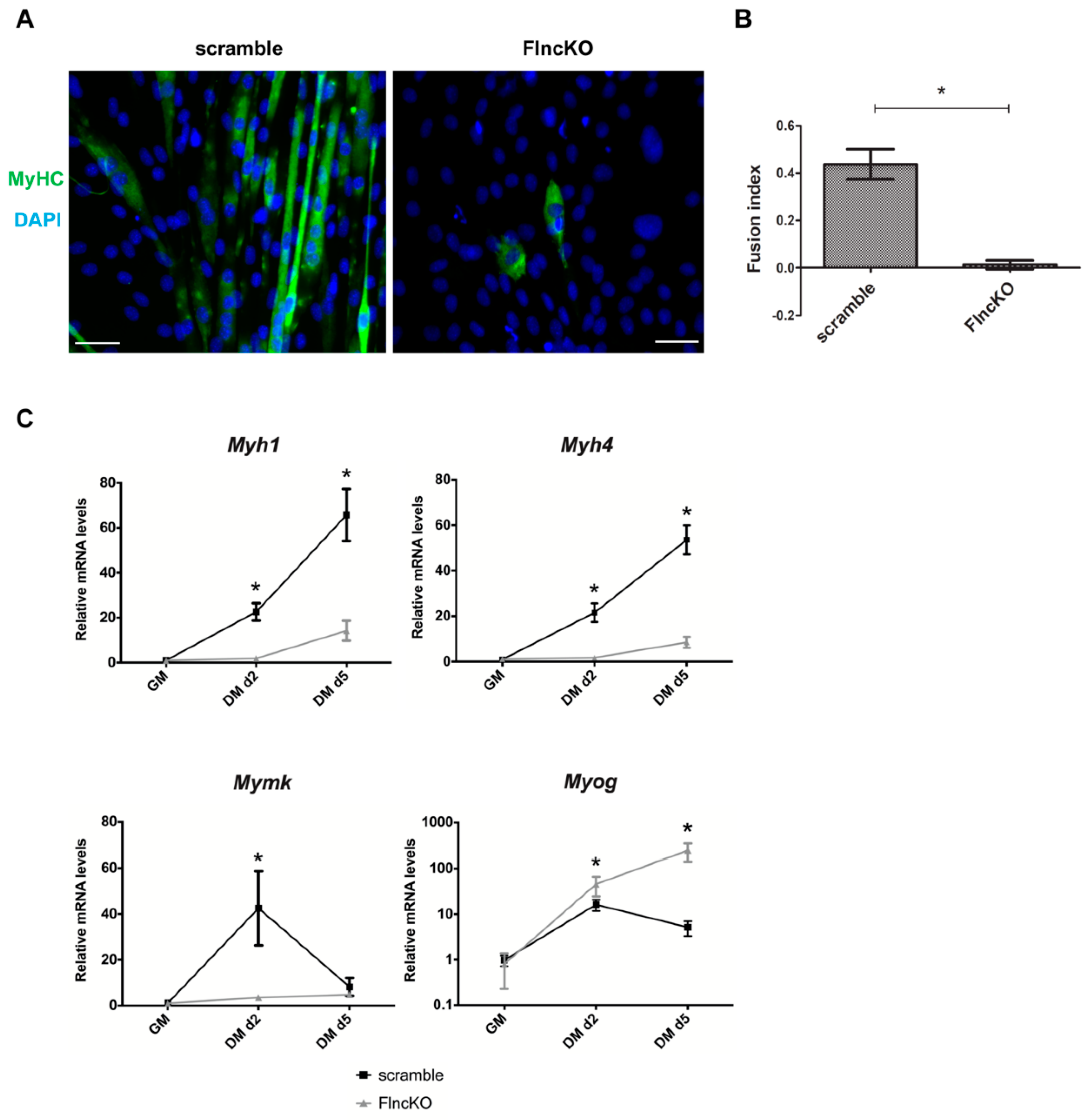
3.3. FlncKO Cells are Characterized by Reduced Ability of Myogenic Differentiation
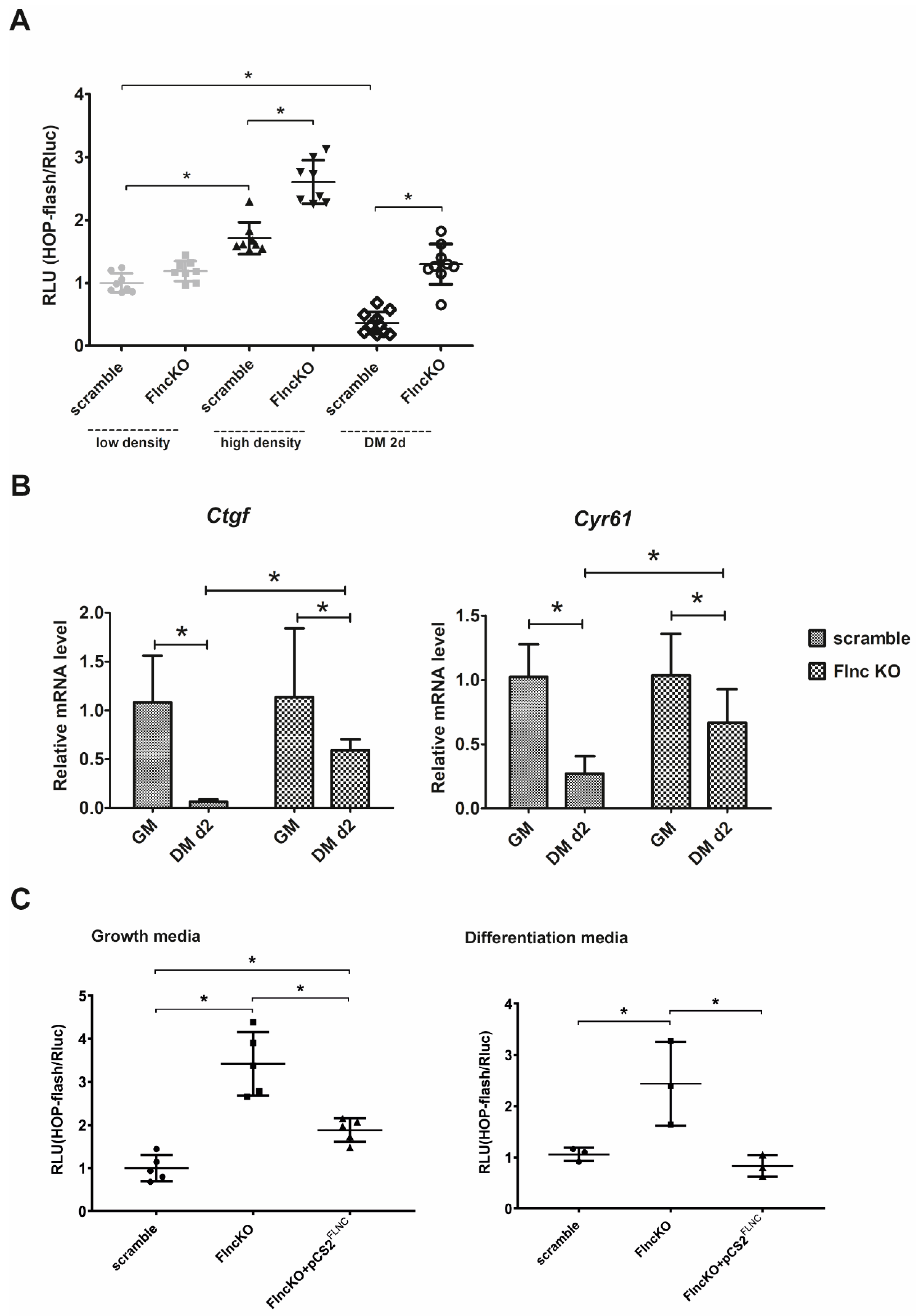
3.4. Downregulation of FLNC in Myoblasts Promotes TEAD-YAP/TAZ Activation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Der Ven, P.F.M.; Obermann, W.M.J.; Lemke, B.; Gautel, M.; Weber, K.; Fürst, D.O. Characterization of Muscle Filamin Isoforms Suggests a Possible Role of γ-Filamin/ABP-L in Sarcomeric Z-Disc Formation. Cell Motil. Cytoskelet. 2000, 45, 149–162. [Google Scholar] [CrossRef]
- Thompson, T.G.; Chan, Y.-M.; Hack, A.A.; Brosius, M.; Rajala, M.; Lidov, H.G.W.; McNally, E.M.; Watkins, S.; Kunkel, L.M. Filamin 2 (FLN2): A Muscle-Specific Sarcoglycan Interacting Protein. J. Cell Biol. 2000, 148, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Kley, R.A.; Hellenbroich, Y.; Van Der Ven, P.F.; Fürst, D.O.; Huebner, A.; Bruchertseifer, V.; Peters, S.A.; Heyer, C.M.; Kirschner, J.; Schröder, R.; et al. Clinical and Morphological Phenotype of the Filamin Myopathy: A Study of 31 German Patients. Brain 2007, 130, 3250–3264. [Google Scholar] [CrossRef] [PubMed]
- Kley, R.A.; Serdaroglu-Oflazer, P.; Leber, Y.; Odgerel, Z.; Van Der Ven, P.F.M.; Olivé, M.; Ferrer, I.; Onipe, A.; Mihaylov, M.; Bilbao, J.M.; et al. Pathophysiology of Protein Aggregation and Extended Phenotyping in Filaminopathy. Brain 2012, 135, 2642–2660. [Google Scholar] [CrossRef]
- Duff, R.M.; Tay, V.; Hackman, P.; Ravenscroft, G.; McLean, C.; Kennedy, P.; Steinbach, A.; Schöffler, W.; Van Der Ven, P.F.M.; Fürst, D.O.; et al. Mutations in the N-Terminal Actin-Binding Domain of Filamin C Cause a Distal Myopathy. Am. J. Hum. Genet. 2011, 88, 729–740. [Google Scholar] [CrossRef]
- Kiselev, A.; Vaz, R.; Knyazeva, A.; Khudiakov, A.; Tarnovskaya, S.; Liu, J.; Sergushichev, A.; Kazakov, S.; Frishman, D.; Smolina, N.; et al. De Novo Mutations in FLNC Leading to Early-Onset Restrictive Cardiomyopathy and Congenital Myopathy. Hum. Mutat. 2018, 39, 1161–1172. [Google Scholar] [CrossRef]
- Ader, F.; De Groote, P.; Réant, P.; Rooryck-Thambo, C.; Dupin-Deguine, D.; Rambaud, C.; Khraiche, D.; Perret, C.; Pruny, J.F.; Mathieu-Dramard, M.; et al. FLNC Pathogenic Variants in Patients with Cardiomyopathies: Prevalence and Genotype-Phenotype Correlations. Clin. Genet. 2019, 96, 317–329. [Google Scholar] [CrossRef]
- van den Bogaart, F.J.A.; Claeys, K.G.; Kley, R.A.; Kusters, B.; Schrading, S.; Kamsteeg, E.J.; Voermans, N.C. Widening the Spectrum of Filamin-C Myopathy: Predominantly Proximal Myopathy Due to the p.A193T Mutation in the Actin-Binding Domain of FLNC. Neuromuscul. Disord. 2017, 27, 73–77. [Google Scholar] [CrossRef]
- Tucker, N.R.; Mclellan, M.A.; Hu, D.; Ye, J.; Parsons, V.A.; Mills, R.W.; Clauss, S.; Dolmatova, E.; Shea, M.A.; Milan, D.J.; et al. Novel Mutation in FLNC (Filamin C) Causes Familial Restrictive Cardiomyopathy. Circ. Cardiovasc. Genet. 2017, 10, e001780. [Google Scholar] [CrossRef]
- Reinstein, E.; Gutierrez-Fernandez, A.; Tzur, S.; Bormans, C.; Marcu, S.; Tayeb-Fligelman, E.; Vinkler, C.; Raas-Rothschild, A.; Irge, D.; Landau, M.; et al. Congenital Dilated Cardiomyopathy Caused by Biallelic Mutations in Filamin C. Eur. J. Hum. Genet. 2016, 24, 1792–1796. [Google Scholar] [CrossRef]
- Ortiz-Genga, M.F.; Cuenca, S.; Dal Ferro, M.; Zorio, E.; Salgado-Aranda, R.; Climent, V.; Padrón-Barthe, L.; Duro-Aguado, I.; Jiménez-Jáimez, J.; Hidalgo-Olivares, V.M.; et al. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J. Am. Coll. Cardiol. 2016, 68, 2440–2451. [Google Scholar] [CrossRef] [PubMed]
- Begay, R.L.; Graw, S.L.; Sinagra, G.; Asimaki, A.; Rowland, T.J.; Slavov, D.B.; Gowan, K.; Jones, K.L.; Brun, F.; Merlo, M.; et al. Filamin C Truncation Mutations Are Associated With Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell–Cell Adhesion Structures. JACC Clin. Electrophysiol. 2018, 4, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Juo, L.-Y.; Liao, W.-C.; Shih, Y.-L.; Yang, B.-Y.; Liu, A.-B.; Yan, Y.-T. HSPB7 Interacts with Dimerized FLNC and Its Absence Results in Progressive Myopathy in Skeletal Muscles. J. Cell Sci. 2016, 129, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Dalkilic, I.; Schienda, J.; Thompson, T.G.; Kunkel, L.M. Loss of FilaminC (FLNc) Results in Severe Defects in Myogenesis and Myotube Structure. Mol. Cell. Biol. 2006, 26, 6522–6534. [Google Scholar] [CrossRef] [PubMed]
- Stossel, T.P.; Condeelis, J.; Cooley, L.; Hartwig, J.H.; Noegel, A.; Schleicher, M.; Shapiro, S.S. Filamins as Integrattors of Cell Mechanics and Signalling. Nat. Rev. Mol. Cell Biol. 2001, 2, 138–145. [Google Scholar] [CrossRef]
- Nakamura, F.; Stossel, T.P.; Hartwig, J.H. The Filamins: Organizers of Cell Structure and Function. Cell Adhes. Migr. 2011, 5, 160–169. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Y.; Zhao, J.; Hei, K.; Zhuang, H.; Li, Q.; Wei, W.; Chen, R.; Zhang, N.; Li, Y. Ectopic Overexpression of Filamin C Scaffolds MEK1/2 and ERK1/2 to Promote the Progression of Human Hepatocellular Carcinoma. Cancer Lett. 2017, 388, 167–176. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in Mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Morikawa, Y.; Heallen, T.; Leach, J.; Xiao, Y.; Martin, J.F. Dystrophin–Glycoprotein Complex Sequesters Yap to Inhibit Cardiomyocyte Proliferation. Nature 2017, 547, 227–231. [Google Scholar] [CrossRef]
- Torrini, C.; Cubero, R.J.; Dirkx, E.; Braga, L.; Ali, H.; Prosdocimo, G.; Gutierrez, M.I.; Collesi, C.; Licastro, D.; Zentilin, L.; et al. Common Regulatory Pathways Mediate Activity of MicroRNAs Inducing Cardiomyocyte Proliferation. Cell Rep. 2019, 27, 2759–2771. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Ruan, G.-X.; Barry, E.; Yu, D.; Lukason, M.; Cheng, S.H.; Scaria, A. CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10. Mol. Ther. 2017, 25, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Rigby, P.W.J. Gene Regulatory Networks and Transcriptional Mechanisms That Control Myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, M.H.; Moskowitz, I.P.; Mendonza, A.M.; Vidali, L.; Nakamura, F.; Kwiatkowski, D.J.; Walsh, C.A. Filamin A (FLNA) Is Required for Cell—Cell Contact in Vascular Development and Cardiac Morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19836–19841. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Jin, T.; Yang, L.; Wei, Q.; Yang, Y.; Li, H.; Zhu, Y. Vinculin and Filamin-C Are Two Potential Prognostic Biomarkers and Therapeutic Targets for Prostate Cancer Cell Migration. Oncotarget 2017, 8, 82430–82436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.-X.; Hartwig, J.H.; Akyürek, L.M. Filamins in Cell Signaling, Transcription and Organ Development. Trends Cell Biol. 2010, 20, 113–123. [Google Scholar] [CrossRef]
- Kamil, M.; Shinsato, Y.; Higa, N.; Hirano, T.; Idogawa, M.; Takajo, T.; Minami, K.; Shimokawa, M.; Yamamoto, M.; Kawahara, K.; et al. High Filamin-C Expression Predicts Enhanced Invasiveness and Poor Outcome in Glioblastoma Multiforme. Br. J. Cancer 2019, 120, 819–826. [Google Scholar] [CrossRef]
- Ulbricht, A.; Eppler, F.J.; Tapia, V.E.; van der Ven, P.F.M.; Hampe, N.; Hersch, N.; Vakeel, P.; Stadel, D.; Haas, A.; Saftig, P.; et al. Cellular Mechanotransduction Relies on Tension-Induced and Chaperone-Assisted Autophagy. Curr. Biol. 2013, 23, 430–435. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, Z.E.; Zhang, L.; Zhu, M.; Tan, C.; Zhou, X.; Evans, S.M.; Fang, X.; Feng, W.; Chen, J. Loss of Filamin C Is Catastrophic for Heart Function. Circulation 2020, 141, 869–871. [Google Scholar] [CrossRef]
- Hall, C.L.; Gurha, P.; Sabater-Molina, M.; Asimaki, A.; Futema, M.; Lovering, R.C.; Suárez, M.P.; Aguilera, B.; Molina, P.; Zorio, E.; et al. RNA Sequencing-Based Transcriptome Profiling of Cardiac Tissue Implicates Novel Putative Disease Mechanisms in FLNC-Associated Arrhythmogenic Cardiomyopathy. Int. J. Cardiol. 2020, 302, 124–130. [Google Scholar] [CrossRef]
- Kostareva, A.; Kiselev, A.; Gudkova, A.; Frishman, G.; Ruepp, A.; Frishman, D.; Smolina, N.; Tarnovskaya, S.; Nilsson, D.; Zlotina, A.; et al. Genetic Spectrum of Idiopathic Restrictive Cardiomyopathy Uncovered by Next-Generation Sequencing. PLoS ONE 2016, 11, e0163362. [Google Scholar] [CrossRef]
- Figeac, N.; Mohamed, A.D.; Sun, C.; Schönfelder, M.; Matallanas, D.; Garcia-Munoz, A.; Missiaglia, E.; Collie-Duguid, E.; De Mello, V.; Pobbati, A.V.; et al. VGLL3 Operates via TEAD1, TEAD3 and TEAD4 to Influence Myogenesis in Skeletal Muscle. J. Cell Sci. 2019, 132, jcs225946. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; De Mello, V.; Mohamed, A.; Ortuste Quiroga, H.P.; Garcia-Munoz, A.; Al Bloshi, A.; Tremblay, A.M.; von Kriegsheim, A.; Collie-Duguid, E.; Vargesson, N.; et al. Common and Distinctive Functions of the Hippo Effectors Taz and Yap in Skeletal Muscle Stem Cell Function. Stem Cells 2017, 35, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.N.; Tremblay, A.M.; Knopp, P.; White, R.B.; Urcia, R.; De Bari, C.; Zammit, P.S.; Camargo, F.D.; Wackerhage, H. The Hippo Pathway Member Yap Plays a Key Role in Influencing Fate Decisions in Muscle Satellite Cells. J. Cell Sci. 2012, 125 Pt 24, 6009–6019. [Google Scholar] [CrossRef]
- Watt, K.I.; Judson, R.; Medlow, P.; Reid, K.; Kurth, T.B.; Burniston, J.G.; Ratkevicius, A.; De Bari, C.; Wackerhage, H. Yap Is a Novel Regulator of C2C12 Myogenesis. Biochem. Biophys. Res. Commun. 2010, 393, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, Y.; Zhang, M.; Heallen, T.; Leach, J.; Tao, G.; Xiao, Y.; Bai, Y.; Li, W.; Willerson, J.T.; Martin, J.F. Actin Cytoskeletal Remodeling with Protrusion Formation Is Essential for Heart Regeneration in Hippo-Deficient Mice. Sci. Signal. 2015, 8, ra41. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knyazeva, A.; Khudiakov, A.; Vaz, R.; Muravyev, A.; Sukhareva, K.; Sejersen, T.; Kostareva, A. FLNC Expression Level Influences the Activity of TEAD-YAP/TAZ Signaling. Genes 2020, 11, 1343. https://doi.org/10.3390/genes11111343
Knyazeva A, Khudiakov A, Vaz R, Muravyev A, Sukhareva K, Sejersen T, Kostareva A. FLNC Expression Level Influences the Activity of TEAD-YAP/TAZ Signaling. Genes. 2020; 11(11):1343. https://doi.org/10.3390/genes11111343
Chicago/Turabian StyleKnyazeva, Anastasia, Aleksandr Khudiakov, Raquel Vaz, Aleksey Muravyev, Ksenia Sukhareva, Thomas Sejersen, and Anna Kostareva. 2020. "FLNC Expression Level Influences the Activity of TEAD-YAP/TAZ Signaling" Genes 11, no. 11: 1343. https://doi.org/10.3390/genes11111343
APA StyleKnyazeva, A., Khudiakov, A., Vaz, R., Muravyev, A., Sukhareva, K., Sejersen, T., & Kostareva, A. (2020). FLNC Expression Level Influences the Activity of TEAD-YAP/TAZ Signaling. Genes, 11(11), 1343. https://doi.org/10.3390/genes11111343




