Evolution of Homeologous Gene Expression in Polyploid Wheat
Abstract
:1. Introduction
2. Materials and Methods
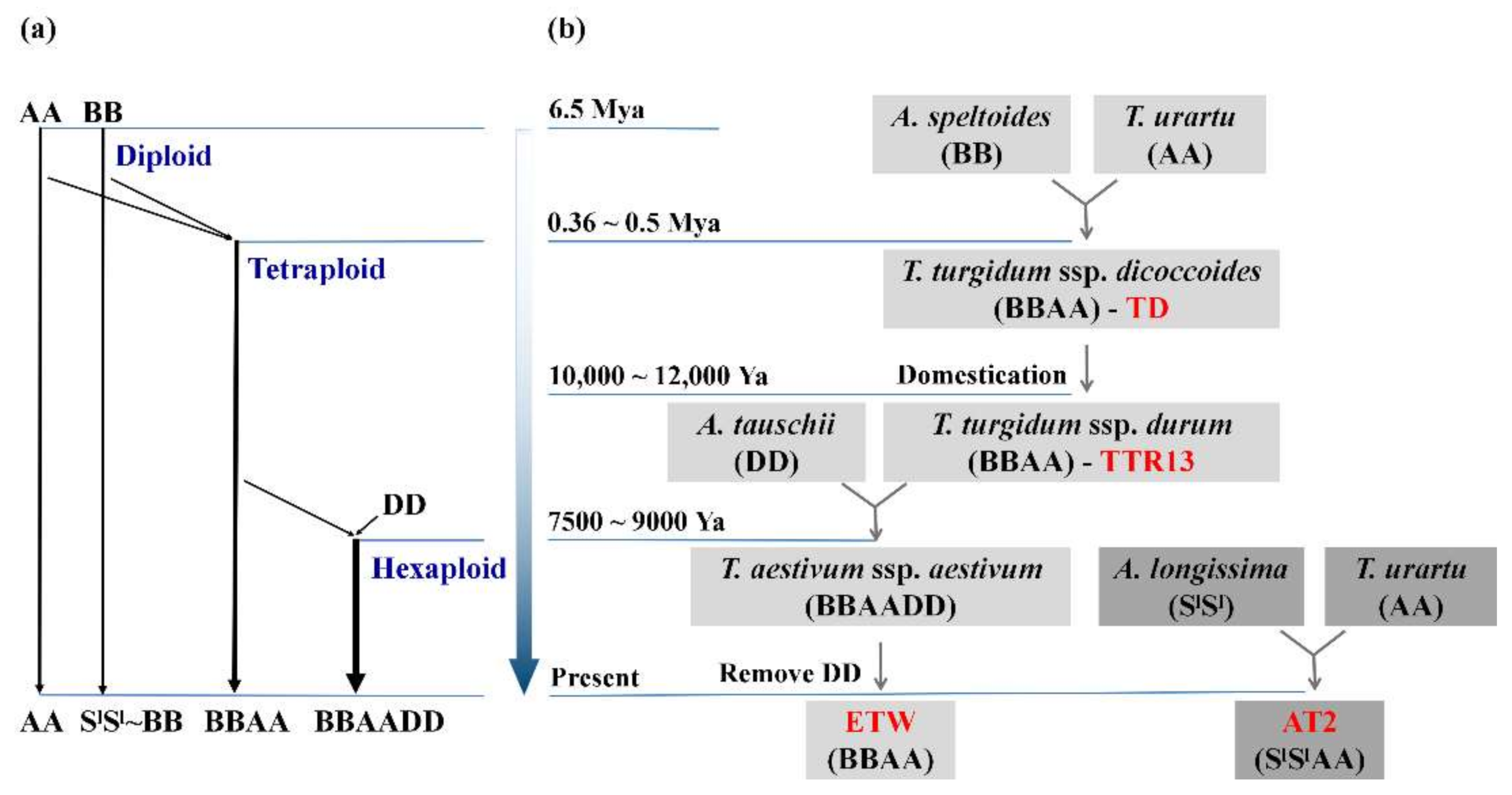
2.1. Plant Materials and RNA-Seq Data
2.2. Preprocessing and Mapping of RNA-Seq Data
2.3. Gene Expression Analysis
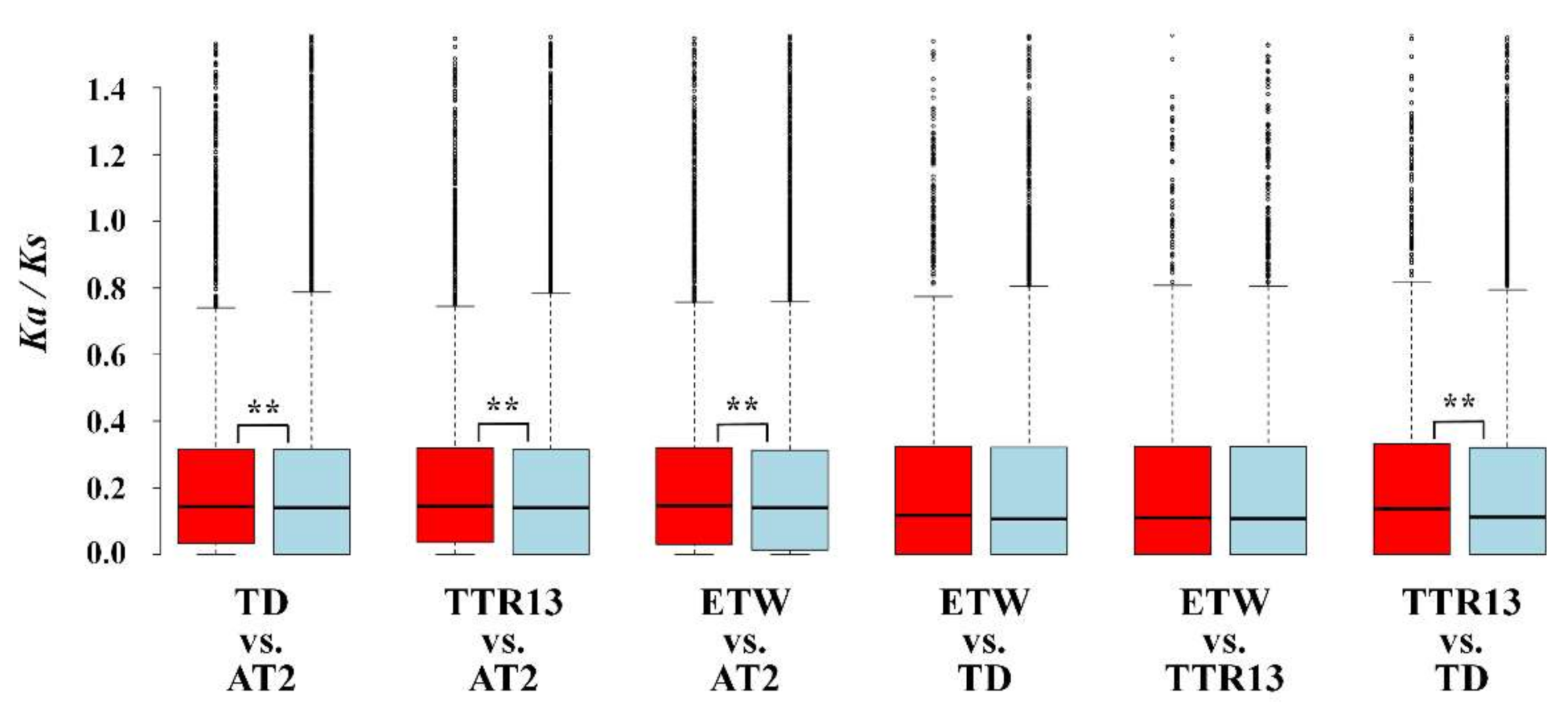
2.4. Calculating Ka/Ks Ratio
2.5. Classification of Genes in Different Duplicate Status within AA or BB Genome
2.6. GO and Pfam Enrichment Analysis
3. Results
3.1. Expression Changes in AA and BB Genomes from Different Ploidy Backgrounds
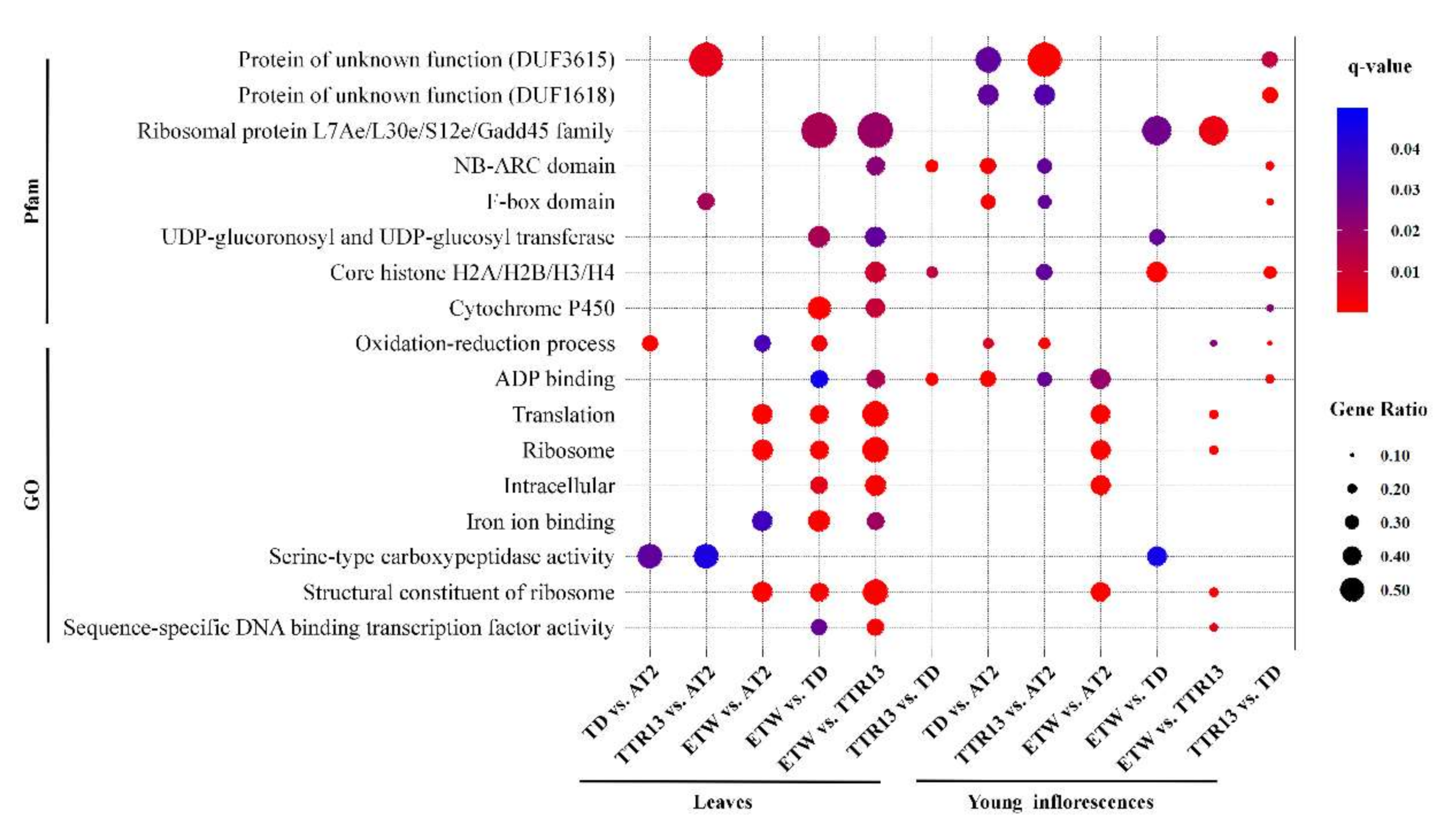
3.2. Enrichment of Gene Functions/Families in DEGs
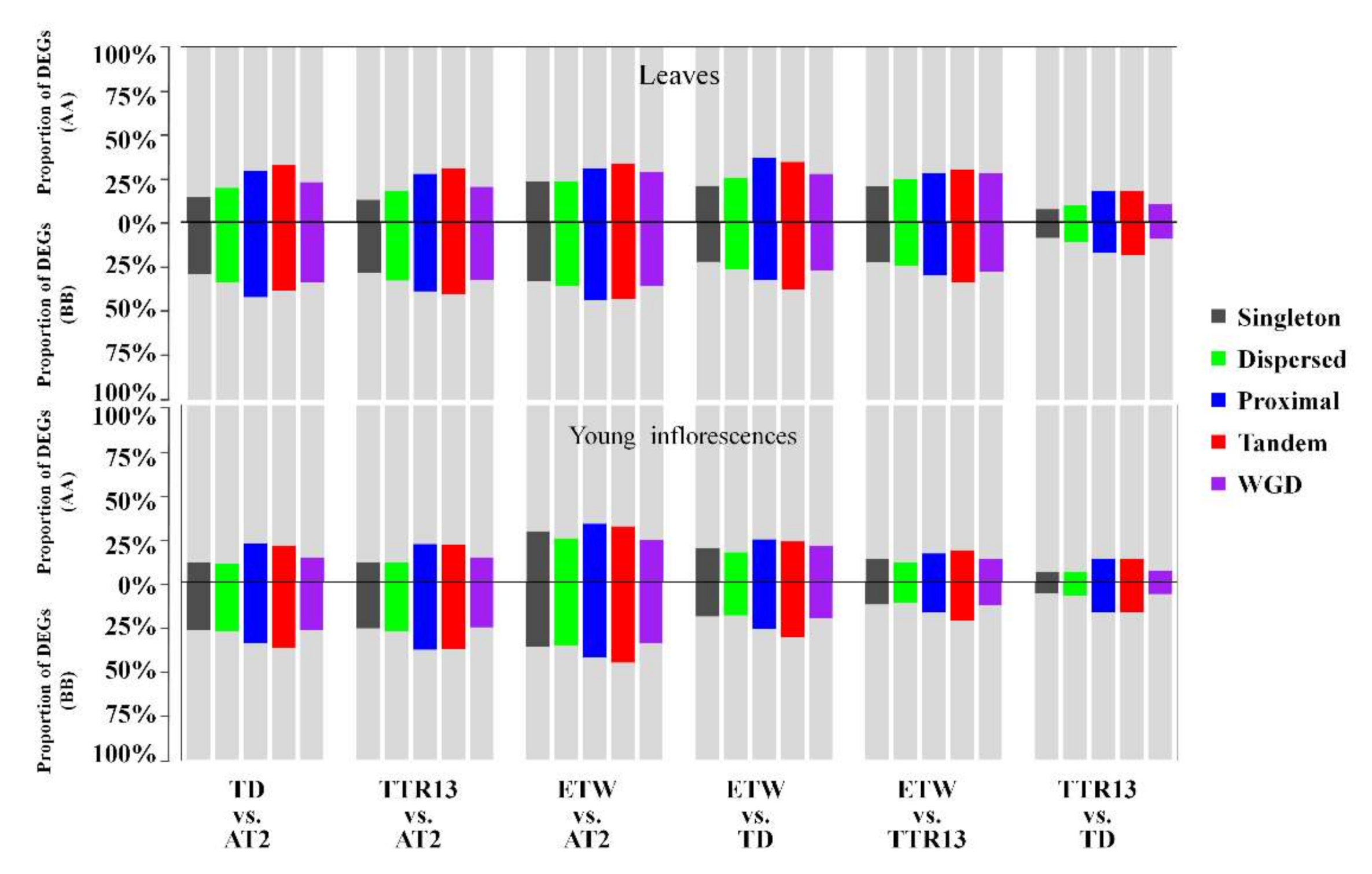
3.3. Distribution of DEGs in Different Duplication Modes
3.4. Distribution of DEGs in A-B Homeolog Pairs
3.5. Relationship between Gene Expression Changes and Selection
4. Discussion
4.1. Effects of Polyploidization on the Evolution of Gene Expression
4.2. Mode of Ancient Gene Duplication Is Related to Gene Expression Changes
4.3. Cis- and Inter-Subgenome Trans-Regulatory Changes Play Roles in the Evolution of Gene Expression in Wheat
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nat. Cell Biol. 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Li, Z.; Defoort, J.; Tasdighian, S.; Maere, S.; Van De Peer, Y.; De Smet, R. Gene Duplicability of Core Genes Is Highly Consistent across All Angiosperms. Plant Cell 2016, 28, 326–344. [Google Scholar] [CrossRef] [Green Version]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome Plasticity a Key Factor in the Success of Polyploid Wheat under Domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Tian, L.; Madlung, A.; Lee, H.-S.; Chen, M.; Lee, J.J.; Watson, B.; Kagochi, T.; Comai, L.; Chen, Z.J. Stochastic and Epigenetic Changes of Gene Expression in Arabidopsis Polyploids. Genetics 2004, 167, 1961–1973. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.A.; Ni, Z.; Scheen, A.-C.; Koh, J.; Gilbert, C.A.; Lefkowitz, D.; Chen, Z.J.; Soltis, P.S.; Soltis, D.E. Evolution and Expression of Homeologous Loci inTragopogon miscellus(Asteraceae), a Recent and Reciprocally Formed Allopolyploid. Genetics 2006, 173, 1599–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, M.-J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Hered. 2013, 110, 171–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Bai, Y.; Lin, X.; Zhao, N.; Hu, L.; Gong, Z.; Wendel, J.F.; Liu, B. Genome-Wide Disruption of Gene Expression in Allopolyploids but Not Hybrids of Rice Subspecies. Mol. Biol. Evol. 2014, 31, 1066–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chagué, V.; Just, J.; Mestiri, I.; Balzergue, S.; Tanguy, A.-M.; Huneau, C.; Huteau, V.; Belcram, H.; Coriton, O.; Jahier, J.; et al. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 2010, 187, 1181–1194. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, B.; Qi, B.; Gou, X.; Dong, Y.; Xu, C.; Zhang, B.; Huang, W.; Liu, C.; Wang, X.; et al. Evolution of the BBAA Component of Bread Wheat during Its History at the Allohexaploid Level. Plant Cell 2014, 26, 2761–2776. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, H.; Li, Y.; Zhang, Z.; Li, L.; Liu, B. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 2015, 209, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Smith, R.; McKain, M.R.; Cooley, A.M.; Vallejo-Marin, M.; Yuan, Y.; Bewick, A.J.; Ji, L.; Platts, A.E.; Bowman, M.J.; et al. Subgenome Dominance in an Interspecific Hybrid, Synthetic Allopolyploid, and a 140-Year-Old Naturally Established Neo-Allopolyploid Monkeyflower. Plant Cell 2017, 29, 2150–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Combes, M.-C.; Alberto, C.; Baraille, H.; Bertrand, B.; Lashermes, P. Homeologous Gene Expression in Response to Growing Temperature in a Recent Allopolyploid (Coffea arabica L.). J. Hered. 2011, 103, 36–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; A Saski, C.; E Scheffler, B.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [Green Version]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.-A.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Dvorak, J.; Akhunov, E.D. Tempos of Gene Locus Deletions and Duplications and Their Relationship to Recombination Rate During Diploid and Polyploid Evolution in the Aegilops-Triticum Alliance. Genetics 2005, 171, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Feldman, M.; Lupton, F.G.H.; Miller, T.E. Wheats. In Evolution of Crop Plants, 2nd ed.; Smartt, J., Simmonds, N.W., Eds.; Longman Scientific: London, UK, 1995; pp. 184–192. [Google Scholar]
- Kerber, E.R. Wheat: Reconstitution of the Tetraploid Component (AABB) of Hexaploids. Science 1964, 143, 253–255. [Google Scholar] [CrossRef]
- Langham, R.J.; Walsh, J.; Dunn, M.; Ko, C.; Goff, S.A.; Freeling, M. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 2004, 166, 935–945. [Google Scholar] [CrossRef]
- De Smet, R.; Adams, K.L.; Vandepoele, K.; Van Montagu, M.C.E.; Maere, S.; Van De Peer, Y. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 2013, 110, 2898–2903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Nadon, B.D.; Kim, K.D.; Jackson, S.A. Genetic and epigenetic divergence of duplicate genes in two legume species. Plant Cell Environ. 2018, 41, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Blanca, G.; Wolfe, K.H. Functional Divergence of Duplicated Genes Formed by Polyploidy during Arabidopsis Evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salse, J.; Bolot, S.; Throude, M.; Jouffe, V.; Piegu, B.; Quraishi, U.M.; Calcagno, T.; Cooke, R.; Delseny, M.; Feuillet, C. Identification and Characterization of Shared Duplications between Rice and Wheat Provide New Insight into Grass Genome Evolution. Plant Cell 2008, 20, 11–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Neph, S.; Kuehn, M.S.; Reynolds, A.P.; Haugen, E.; Thurman, R.E.; Johnson, A.K.; Rynes, E.; Maurano, M.T.; Vierstra, J.; Thomas, S.; et al. BEDOPS: High-performance genomic feature operations. Bioinformatics 2012, 28, 1919–1920. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 002832. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Stajich, J.; Block, D.; Boulez, K.; Brenner, S.E.; Chervitz, S.A.; Dagdigian, C.; Fuellen, G.; Gilbert, J.G.; Korf, I.; Lapp, H.; et al. The Bioperl Toolkit: Perl Modules for the Life Sciences. Genome Res. 2002, 12, 1611–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Wendel, J. Genetic and Epigenetic Aspects of Polyploid Evolution in Plants. Cytogenet. Genome Res. 2013, 140, 270–285. [Google Scholar] [CrossRef]
- Liu, B.; Vega, J.; Feldman, M. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 1998, 41, 535–542. [Google Scholar] [CrossRef]
- Ozkan, H.; Levy, A.A.; Feldman, M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 2001, 13, 1735–1747. [Google Scholar]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nat. Cell Biol. 2012, 491, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Saintenac, C.; Jiang, D.; Akhunov, E.D. Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 2011, 12, R88-17. [Google Scholar] [CrossRef] [Green Version]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, N.; Zhu, B.; Li, M.; Wang, L.; Xu, L.; Zhang, H.; Zheng, S.; Qi, B.; Han, F.; Liu, B. Extensive and Heritable Epigenetic Remodeling and Genetic Stability Accompany Allohexaploidization of Wheat. Genetics 2011, 188, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Shitsukawa, N.; Tahira, C.; Kassai, K.-I.; Hirabayashi, C.; Shimizu, T.; Takumi, S.; Mochida, K.; Kawaura, K.; Ogihara, Y.; Murai, K. Genetic and Epigenetic Alteration among Three Homoeologous Genes of a Class E MADS Box Gene in Hexaploid Wheat. Plant Cell 2007, 19, 1723–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970. [Google Scholar]
- Duarte, J.M.; Wall, P.K.; Edger, P.P.; Landherr, L.L.; Ma, H.; Pires, J.C.; Leebens-Mack, J.; Depamphilis, C.W. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 2010, 10, 61. [Google Scholar] [CrossRef] [Green Version]
- Birchler, J.A.; Bhadra, U.; Pal-Bhadra, M.; Auger, D.L. Dosage-Dependent Gene Regulation in Multicellular Eukaryotes: Implications for Dosage Compensation, Aneuploid Syndromes, and Quantitative Traits. Dev. Biol. 2001, 234, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosom. Res. 2009, 17, 699–717. [Google Scholar] [CrossRef] [Green Version]
- Tasdighian, S.; Van Bel, M.; Li, Z.; Van De Peer, Y.; Carretero-Paulet, L.; Maere, S. Reciprocally Retained Genes in the Angiosperm Lineage Show the Hallmarks of Dosage Balance Sensitivity. Plant Cell 2017, 29, 2766–2785. [Google Scholar] [CrossRef] [Green Version]
- Teichmann, S.A.; Babu, M.M. Gene regulatory network growth by duplication. Nat. Genet. 2004, 36, 492–496. [Google Scholar] [CrossRef]
- Gu, X.; Zhang, Z.; Huang, W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc. Natl. Acad. Sci. USA 2005, 102, 707–712. [Google Scholar] [CrossRef] [Green Version]
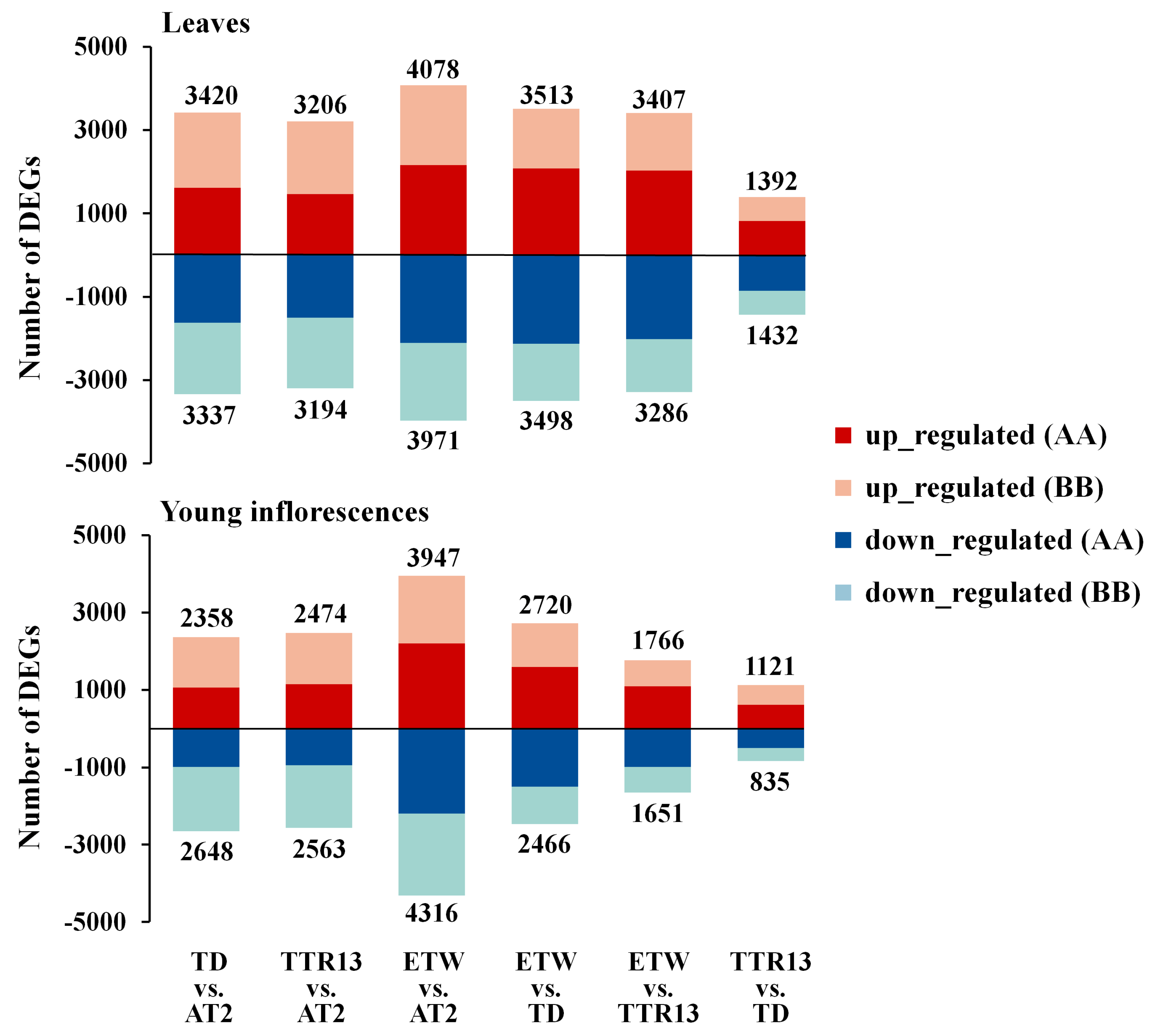
| TD vs. AT2 | TTR13 vs. AT2 | ETW vs. AT2 | ETW vs. TD | ETW vs. TTR13 | TTR13 vs. TD | |
|---|---|---|---|---|---|---|
| Leaves a | 4418 | 4136 | 5360 | 4694 | 4651 | 1496 |
| DEG in A or B b | 3839 | 3646 | 4502 | 3788 | 3715 | 1363 |
| A-DEG | 1312 | 1206 | 1806 | 1841 | 1817 | 620 |
| B-DEG | 2527 | 2440 | 2696 | 1947 | 1898 | 743 |
| DEG in A and B c | 579 | 490 | 858 | 906 | 936 | 133 |
| AB-convergent | 416 | 330 | 671 | 846 | 892 | 105 |
| AB-divergent | 163 | 160 | 187 | 60 | 44 | 28 |
| Young inflorescences a | 2866 | 2944 | 5108 | 3071 | 1873 | 968 |
| DEG in A or B b | 2621 | 2675 | 4352 | 2543 | 1598 | 879 |
| A-DEG | 629 | 645 | 1648 | 1169 | 730 | 372 |
| B-DEG | 1992 | 2030 | 2704 | 1374 | 868 | 507 |
| DEG in A and B c | 245 | 269 | 756 | 528 | 275 | 89 |
| AB-convergent | 138 | 151 | 588 | 494 | 259 | 69 |
| AB-divergent | 107 | 118 | 168 | 34 | 16 | 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Dong, Q.; Nadon, B.D.; Ding, X.; Wang, X.; Dong, Y.; Liu, B.; Jackson, S.A.; Xu, C. Evolution of Homeologous Gene Expression in Polyploid Wheat. Genes 2020, 11, 1401. https://doi.org/10.3390/genes11121401
Zhao N, Dong Q, Nadon BD, Ding X, Wang X, Dong Y, Liu B, Jackson SA, Xu C. Evolution of Homeologous Gene Expression in Polyploid Wheat. Genes. 2020; 11(12):1401. https://doi.org/10.3390/genes11121401
Chicago/Turabian StyleZhao, Na, Qianli Dong, Brian D. Nadon, Xiaoyang Ding, Xutong Wang, Yuzhu Dong, Bao Liu, Scott A. Jackson, and Chunming Xu. 2020. "Evolution of Homeologous Gene Expression in Polyploid Wheat" Genes 11, no. 12: 1401. https://doi.org/10.3390/genes11121401





