The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. BAC Clone DNA Isolation and Labeling
2.3. Oligonucleotide Probe Design, Synthesis, and Labeling
2.4. Fluorescence In Situ Hybridization
3. Results
Oligonucleotide-Based Probe Development and Oligo-FISH
- The chromosome region should exhibit at least partial differentiation in related species, evidenced by previous cytogenetic studies or genome/linkage mapping;
- The chromosome region should have a low abundance of repetitive elements to allow the design of unique probes;
- Scaffolding in this region should be strongly supported by linkage mapping to avoid unintentional incorporation of fragments from other chromosomes;
- Chromosome-specific cytogenetic landmarks (i.e., BAC clones) should be available for this region to enable parallel use of two techniques—BAC-FISH and oligo-FISH.
4. Discussion
4.1. Development of Oligonucleotide Probe Sets
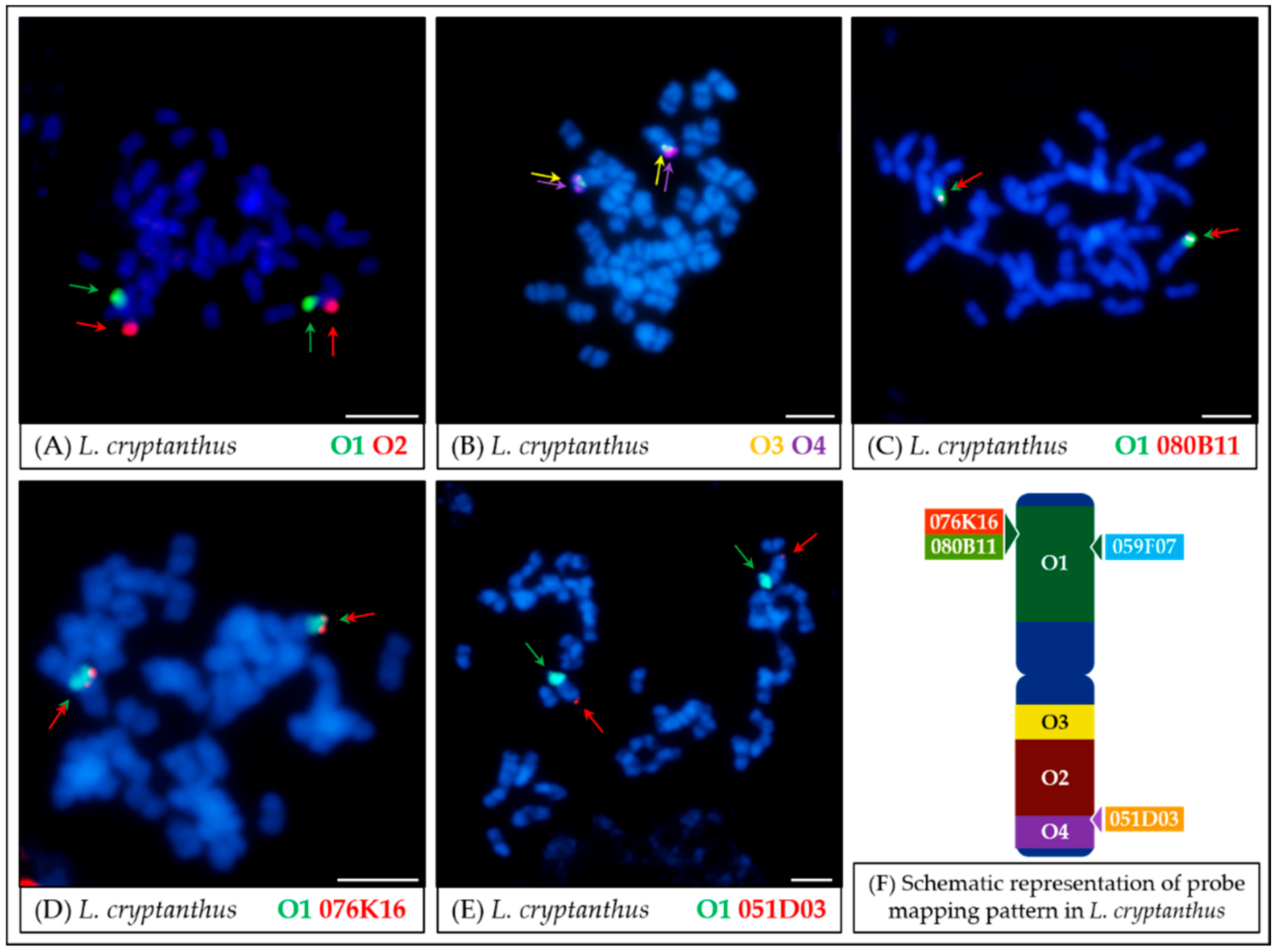
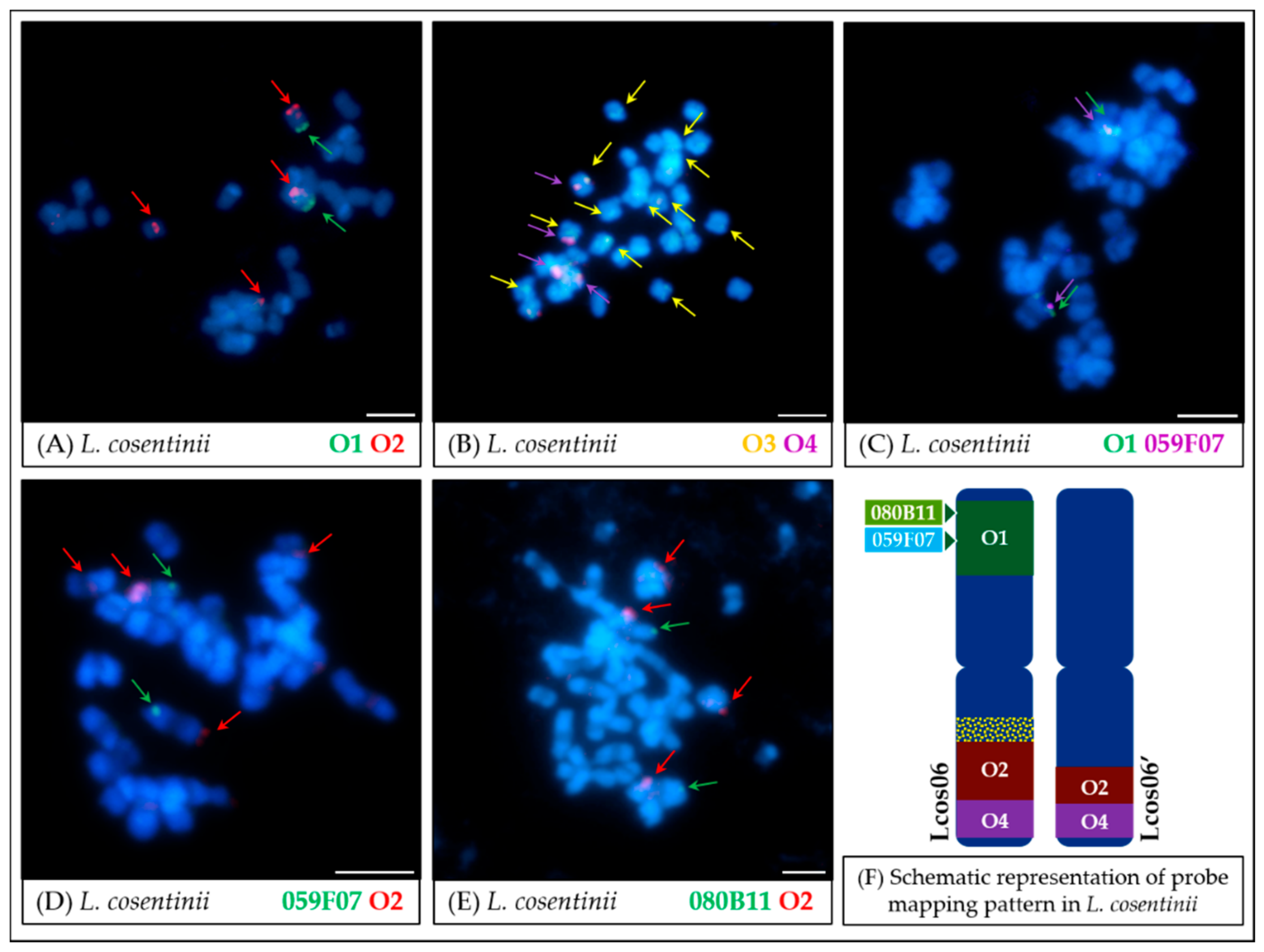
4.2. Comparative Mapping of Wild Lupin Species Using Oligonucleotide Probes
5. Conclusions
- In the case of L. micranthus (Figure 5 and Figure 8), evolutionally the oldest among the studied species, the probes specific to the Lang06 chromosome landed on two different chromosomes, which may represent the pattern in the common ancestor of Old World lupins. During the course of evolution and speciation, the two genome fragments were translocated to one chromosome.
- In L. cosentinii (Figure 6 and Figure 8), hybridization of both O2 and O4 to two different chromosomes, as well as the highest number of probes (BACs and oligonucleotides) dispersed on multiple loci, might be the result of duplication and/or translocation of the arm B fragment of Lang06 chromosome (containing O1 and O2 probes) to Lcos06′ chromosome.
- Significant synteny changes detected in L. pilosus (Figure 7 and Figure 8) were probably the result of a series of rearrangements, including translocation, paracentric inversion, and/or non-allelic homologous recombination, leading to the separation of probes derived from Lang06 into three individual L. pilosus chromosomes.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gepts, P.; Beavis, W.D.; Brummer, E.C.; Shoemaker, R.C.; Stalker, H.T.; Weeden, N.F.; Young, N.D. Legumes as a model plant family. genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 2005, 137, 1228–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, J.J.; Luckow, M.A. The rest of the iceberg. legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003, 131, 900–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, R.; Lewis, G.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the world. TAXON 2006, 55, 251. [Google Scholar] [CrossRef]
- Bertioli, D.; Moretzsohn, M.C.; Madsen, L.H.; Sandal, N.; Leal-Bertioli, S.C.M.; Guimarães, P.M.; Hougaard, B.K.; Fredslund, J.; Schauser, L.; Nielsen, A.M.; et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genom. 2009, 10, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.A.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, D.E.; Visger, C.J.; Marchant, D.B.; Soltis, P.S. Polyploidy: Pitfalls and paths to a paradigm. Am. J. Bot. 2016, 103, 1146–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; McKain, M.R.; Harkess, A.; Nelson, M.N.; Dash, S.; Deyholos, M.K.; Peng, Y.; Joyce, B.; Stewart, C.N.; Rolf, M.; et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol. Biol. Evol. 2015, 32, 193–210. [Google Scholar] [CrossRef]
- Drummond, C.S.; Eastwood, R.J.; Miotto, S.T.S.; Hughes, C.E. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovation with incomplete Taxon sampling. Syst. Biol. 2012, 61, 443–460. [Google Scholar] [CrossRef] [Green Version]
- LPWG. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). TAXON 2017, 66, 44–77. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Zhang, X.; Qi, J. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Petterson, D.S. The use of Lupins in feeding systems—Review. Asian-Australas. J. Anim. Sci. 2000, 13, 861–882. [Google Scholar] [CrossRef]
- Lucas, M.M.; Stoddard, F.L.; Annicchiarico, P.; Frias, J.; Emartinez-Villaluenga, C.; Esussmann, D.; Duranti, M.M.; Eseger, A.; Zander, P.; Pueyo, J. The future of lupin as a protein crop in Europe. Front. Plant Sci. 2015, 6, 705. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, Q.; Yuan, W.; Xu, F.; Aslam, M.M.; Miao, R.; Li, Y.; Wang, Q.; Li, X.; Zhang, X.; et al. The genome evolution and low-phosphorus adaptation in white lupin. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, M.M.; Erskine, W.; Berger, J.D.; Udall, J.A.; Nelson, M.N. Genomics of Yellow Lupin (Lupinus luteus L.); Springer Science and Business Media LLC: Berlin, Germany, 2020; pp. 151–159. [Google Scholar]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Doumas, P.; Sallet, E.; Mancinotti, D.; Carrère, S.; Marande, W.; et al. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyrwa, K.; Książkiewicz, M.; Koczyk, G.; Szczepaniak, A.; Podkowiński, J.; Naganowska, B. A tale of two families: Whole genome and segmental duplications underlie glutamine synthetase and phosphoenolpyruvate carboxylase diversity in narrow-leafed Lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 2020, 21, 2580. [Google Scholar] [CrossRef] [Green Version]
- Susek, K.; Bielski, W.; Wyrwa, K.; Hasterok, R.; Jackson, S.A.; Wolko, B.; Naganowska, B. Impact of chromosomal rearrangements on the interpretation of Lupin Karyotype evolution. Genes 2019, 10, 259. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Jian, J.; Wang, P.; Li, C.; Tao, Y.; Li, X.; Renshaw, D.; Clements, J.; Sweetingham, M.; Yang, H. Construction of an ultra-high density consensus genetic map, and enhancement of the physical map from genome sequencing in Lupinus angustifolius. Theor. Appl. Genet. 2018, 131, 209–223. [Google Scholar] [CrossRef]
- Gladstones, J. Distribution, origin, taxonomy, history and importance. In Lupins as Crop Plants: Biology, Production, and Utilization; Gladstones, J.S., Atkins, C.A., Hamblin, J., Eds.; CAB International: Wallingford, CT, USA, 1998; pp. 1–36. [Google Scholar]
- Pazy, B.; Heyn, C.; Herrnstadt, I.; Plitmann, U. Studies in populations of the old world Lupinus species. I. chromosomes of the East Mediterranean lupines. Isr. J. Bot. 1977. [Google Scholar]
- Naganowska, B.; Wolko, B.; Śliwińska, E.; Kaczmarek, Z. Nuclear DNA content variation and species relationships in the genus Lupinus (Fabaceae). Ann. Bot. 2003, 92, 349–355. [Google Scholar] [CrossRef]
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef]
- Plitmann, U.; Pazy, B. Cytogeographical distribution of the Old World Lupinus. Webbia 1984, 38, 531–540. [Google Scholar] [CrossRef]
- Gladstones, J. Lupins of the Mediterranean Region and Africa; Western Australian Department of Agriculture: Kensington, Australia, 1974. [Google Scholar]
- Mahé, F.; Pascual, H.; Coriton, O.; Huteau, V.; Perris, A.N.; Misset, M.-T.; Ainouche, A. New data and phylogenetic placement of the enigmatic Old World lupin: Lupinus mariae-josephi H. Pascual. Genet. Resour. Crop. Evol. 2010, 58, 101–114. [Google Scholar] [CrossRef]
- Jiang, J.; Gill, B.S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 2006, 49, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Langer-Safer, P.R.; Levine, M.; Ward, D.C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 1982, 79, 4381–4385. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosom. Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef] [Green Version]
- Qu, M.; Li, K.; Han, Y.; Chen, L.; Li, Z.; Han, Y. Integrated karyotyping of Woodland Strawberry (Fragaria vesca) with Oligopaint FISH probes. Cytogenet. Genome Res. 2017, 153, 158–164. [Google Scholar] [CrossRef]
- Braz, G.T.; He, L.; Zhao, H.; Marand, A.P.; Semrau, K.; Rouillard, J.-M.; Torres, G.A.; Jiang, J. Comparative Oligo-FISH mapping: An efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2017, 208, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Šimoníková, D.; Němečková, A.; Karafiátová, M.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa Spp.). Front. Plant. Sci. 2019, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Kasprzak, A.; Šafář, J.; Janda, J.; Doležel, J.; Wolko, B.; Naganowska, B. The bacterial artificial chromosome (BAC) library of the narrow-leafed lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 2006, 11, 396–407. [Google Scholar] [CrossRef]
- Wyrwa, K.; Książkiewicz, M.; Szczepaniak, A.; Susek, K.; Podkowiński, J.; Naganowska, B. Integration of Lupinus angustifolius L. (narrow-leafed lupin) genome maps and comparative mapping within legumes. Chromosom. Res. 2016, 24, 355–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Susek, K.; Bielski, W.; Hasterok, R.; Naganowska, B.; Wolko, B. A first glimpse of Wild Lupin karyotype variation as revealed by comparative cytogenetic mapping. Front. Plant. Sci. 2016, 7, 1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Książkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.N.; Renshaw, D.; Rychel, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Hane, J.K.; Ming, Y.; Kamphuis, L.G.; Nelson, M.N.; Garg, G.; Atkins, C.A.; Bayer, P.E.; Bravo, A.; Bringans, S.; Cannon, S.; et al. A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: Insights into plant-microbe interactions and legume evolution. Plant. Biotechnol. J. 2016, 15, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 8 December 2020).
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef] [Green Version]
- Kamphuis, L.G.; Hane, J.K.; Nelson, M.N.; Gao, L.; Atkins, C.A.; Singh, K.B. Transcriptome sequencing of different narrow-leafed lupin tissue types provides a comprehensive uni-gene assembly and extensive gene-based molecular markers. Plant. Biotechnol. J. 2015, 13, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Murgha, Y.E.; Rouillard, J.-M.; Gulari, E. Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS ONE 2014, 9, e94752. [Google Scholar] [CrossRef] [Green Version]
- Beliveau, B.; Joyce, E.; Apostolopoulos, N.; Yilmaz, F.; Fonseka, C.; McCole, R.; Chang, Y.; Li, J.; Senaratne, T.; Williams, B. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 2012, 109, 21301–21306. [Google Scholar] [CrossRef] [Green Version]
- Eastwood, R.; Drummond, C.; Schifino-Wittmann, M.; Hughes, C. Diversity and Evolutionary History of Lupins-Insights from New Phylogenies. Lupins for Health and Wealth; International Lupin Association: Canterbury, New Zealand, 2008; pp. 346–354. [Google Scholar]
- Albert, P.S.; Marand, A.P.; Semrau, K.; Rouillard, J.-M.; Kao, Y.-H.; Wang, C.-J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Gill, B.S. Nonisotopic in situ hybridization and plant genome mapping: The first 10 years. Genome 1994, 37, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Książkiewicz, M.; Rychel, S.; Nelson, M.N.; Wyrwa, K.; Naganowska, B.; Wolko, B. Expansion of the phosphatidylethanolamine binding protein family in legumes: A case study of Lupinus angustifolius L. Flowering Locus T homologs, LanFTc1 and LanFTc2. BMC Genom. 2016, 17, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, M.N.; Książkiewicz, M.; Rychel, S.; Besharat, N.; Taylor, C.M.; Wyrwa, K.; Jost, R.; Erskine, W.; Cowling, W.A.; Berger, J.; et al. The loss of vernalization requirement in narrow-leafed lupin is associated with a deletion in the promoter and de-repressed expression of a Flowering Locus T(FT) homologue. New Phytol. 2016, 213, 220–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Group | Section | Species | Accession | Chromosome Number (2n) | Genome Size (pg/2C DNA) |
|---|---|---|---|---|---|
| Smooth-seeded | Angustifolius | L. angustifolius L. | cv. ‘Sonet’ | 40 | 1.89 |
| L. cryptanthus Shuttlew | 96361 | 40 | 1.86 | ||
| Micranthus | L. micranthus Guss. | 98552 | 52 | 0.98 | |
| Rough-seeded | Pilosus | L. cosentinii Guss. | 98452 | 32 | 1.42 |
| L. pilosus Murr. | 98653 | 42 | 1.36 |
| Oligo Library ID | Lang06 arm (A/B) | Number of Oligonucleotides in Pool | Covered Region in Template Sequence (bp) | Template Coverage | Average Density |
|---|---|---|---|---|---|
| O1 | A | 20,115 | 32,610–11,689,408 | 28.50% | 0.58 oligo/kb |
| O2 | B | 19,926 | 29,400,911–40,902,159 | 28.19% | 0.58 oligo/kb |
| O3 | B | 10,214 | 27,898,181–36,244,755 | 20.41% | 0.82 oligo/kb |
| O4 | B | 8001 | 38,482,724–40,902,115 | 5.91% | 0.30 oligo/kb |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bielski, W.; Książkiewicz, M.; Šimoníková, D.; Hřibová, E.; Susek, K.; Naganowska, B. The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping. Genes 2020, 11, 1489. https://doi.org/10.3390/genes11121489
Bielski W, Książkiewicz M, Šimoníková D, Hřibová E, Susek K, Naganowska B. The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping. Genes. 2020; 11(12):1489. https://doi.org/10.3390/genes11121489
Chicago/Turabian StyleBielski, Wojciech, Michał Książkiewicz, Denisa Šimoníková, Eva Hřibová, Karolina Susek, and Barbara Naganowska. 2020. "The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping" Genes 11, no. 12: 1489. https://doi.org/10.3390/genes11121489
APA StyleBielski, W., Książkiewicz, M., Šimoníková, D., Hřibová, E., Susek, K., & Naganowska, B. (2020). The Puzzling Fate of a Lupin Chromosome Revealed by Reciprocal Oligo-FISH and BAC-FISH Mapping. Genes, 11(12), 1489. https://doi.org/10.3390/genes11121489





