The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Identification and Sequence Analysis of RALF-Like Proteins
2.3. Maximum Likelihood Phylogeny of RALF-Like Proteins
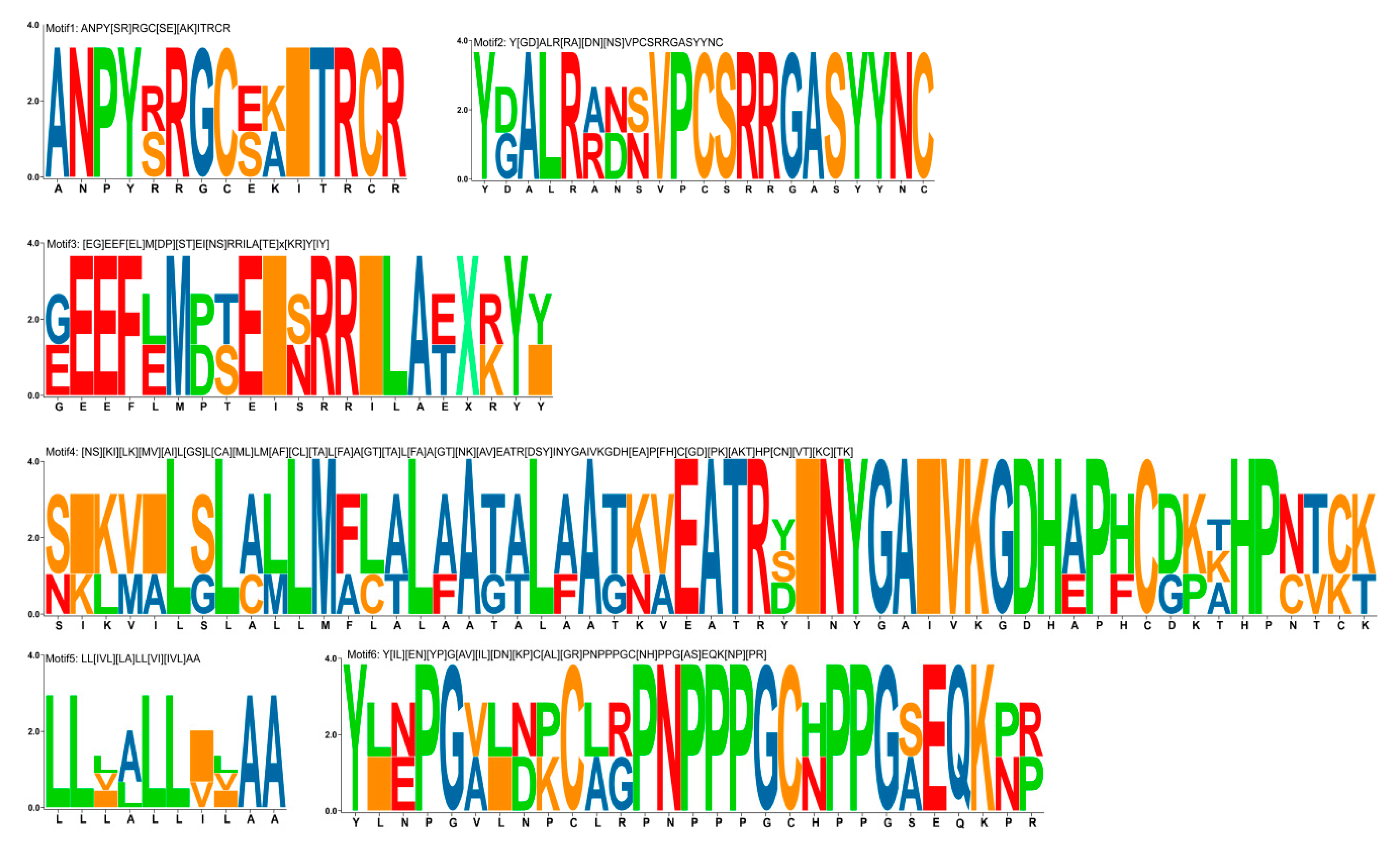
2.4. The Conserved Domain and Motif Analysis
2.5. The Coding Capacity of RALF-Like Genes in the Seven Rosaceae Species
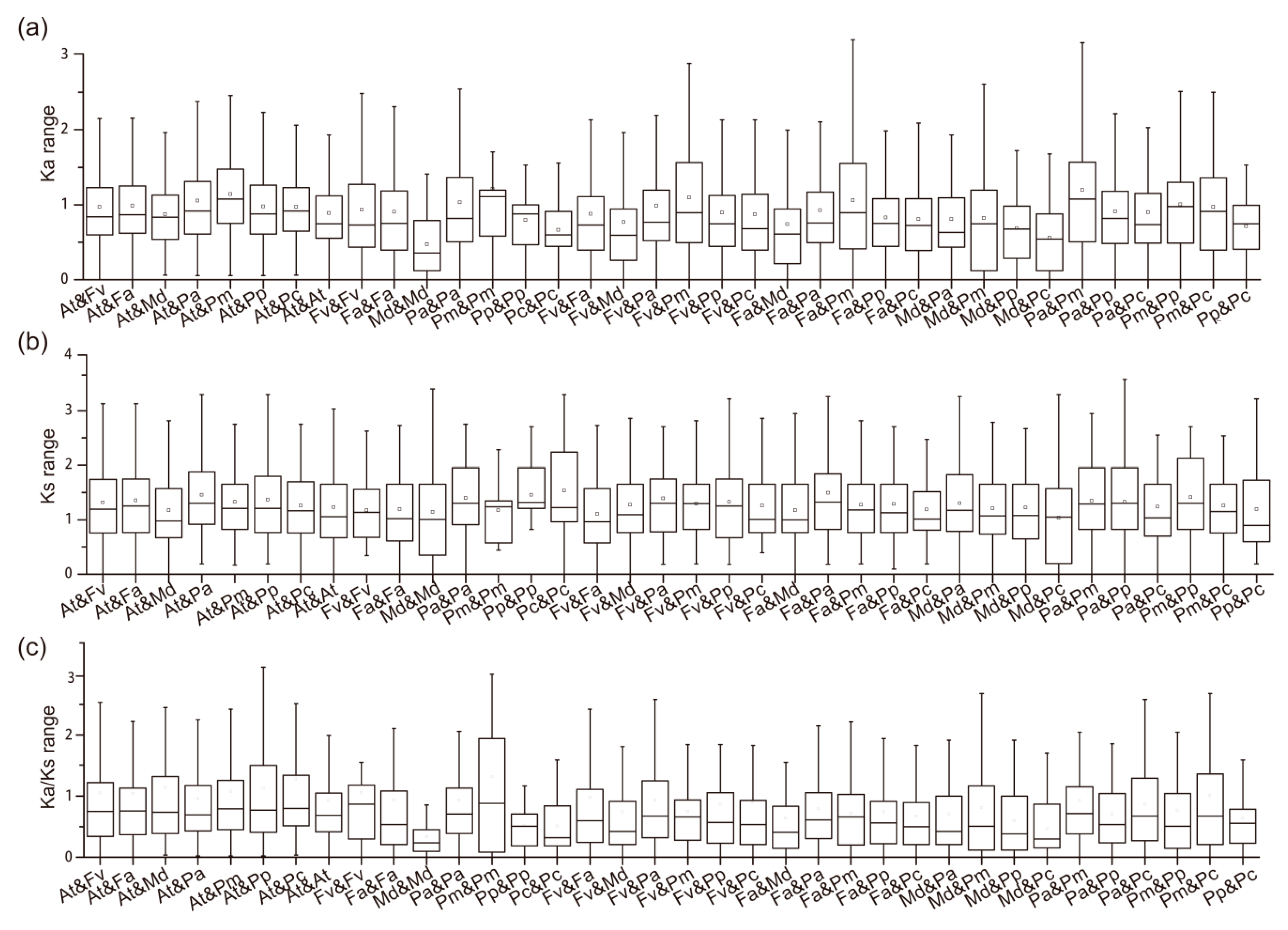
2.6. Duplication Event Analysis and ka and ks Values among RALF-Like Genes
2.7. The Expression Analysis Using Expression Sequence Tag Data
2.8. Gene Ontology and cis-Regulatory Elements Analysis
2.9. The Expression Profiles of RALF-Like Genes in Strawberry and Apple from RNA-Sequencing
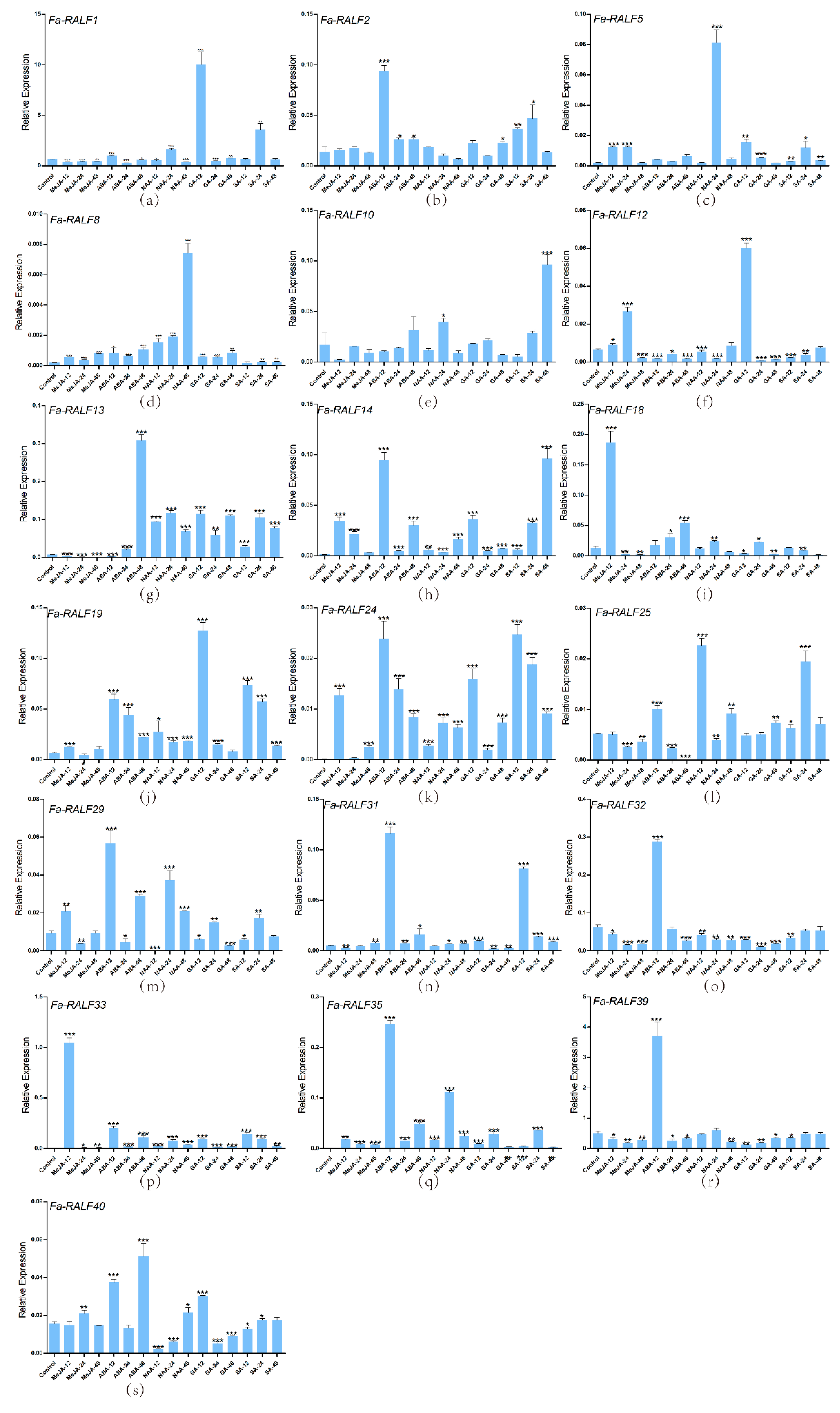
2.10. Gene Validation Experiments by Quantitative Real-Time PCR (RT-qPCR)
3. Results
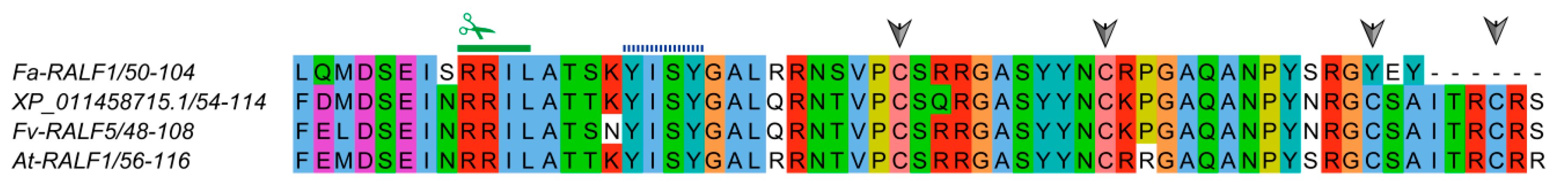
3.1. Identification, Coding Capacity, Classification, and Structural Features of RALF-Like Genes in Rosaceae Species
3.2. Chromosome Location, Duplication Events, and Divergence Rates
3.3. Gene Ontology and Cis-Regulatory Elements Analysis
3.4. The Expression Information from Expression Sequence Tag Data
3.5. The RNA-Sequencing Expression of RALF-Like Genes in Strawberry and Apple
3.6. RT-qPCR Analysis of Nineteen RALF-Like Genes in Cultivated Strawberry Related to Hormones
4. Discussion
4.1. Expression Profile of RALF-Like Genes in Seven Rosaceae Species
4.2. Classification, Duplication Events and Evolution of RALF-Like Proteins in Seven Rosaceae Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Murphy, E.; Smet, I.D. Understanding the RALF family: A tale of many species. Trends Plant Sci. 2014, 19, 664–671. [Google Scholar] [CrossRef]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A., Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar] [CrossRef] [Green Version]
- Bedinger, P.A.; Pearce, G.; Covey, P.A. RALFs: Peptide regulators of plant growth. Plant Signal. Behav. 2010, 5, 1342–1346. [Google Scholar] [CrossRef] [Green Version]
- Campbell, L.; Turner, S.R. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 2017, 8, 37. [Google Scholar] [CrossRef]
- Haruta, M.; Constabel, C.P. Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning, and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiol. 2003, 131, 814–823. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.N.; Mundy, J.; Skriver, K. Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. Silico Biol. 2002, 2, 441–451. [Google Scholar]
- Germain, H.; Chevalier, É.; Caron, S.; Matton, D.P. Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 2005, 220, 447–454. [Google Scholar] [CrossRef]
- Silverstein, K.A.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Cao, J.; Shi, F. Evolution of the RALF gene family in plants: Gene duplication and selection patterns. Evol. Bioinform. 2012, 8, 271–292. [Google Scholar] [CrossRef]
- Ghorbani, S.; Lin, Y.C.; Parizot, B.; Fernandez, A.; Njo, M.F.; Peer, Y.V.D.; Beeckman, T.; Hilson, P. Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J. Exp. Bot. 2015, 66, 5257–5269. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Hussain, A.; Mun, B.G.; Imran, Q.M.; Falak, N.; Lee, S.U.; Kim, J.Y.; Hong, J.K.; Loake, G.J.; Ali, A.; et al. Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 2016, 160, 82–90. [Google Scholar] [CrossRef]
- Macintosh, G.C.; Wilkerson, C.; Green, P.J. Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol. 2001, 127, 765–776. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P.; et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef] [Green Version]
- Ge, A.X.; Zhao, Y.L.; Liu, M.C.; Zhou, L.Z.; Wang, L.L.; Zhong, S.; Hou, S.Y.; Jiang, J.H.; Liu, T.X.; Huang, Q.P.; et al. LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr. Biol. 2019, 29, 3256–3265. [Google Scholar] [CrossRef]
- Moussu, S.; Broyart, C.; Santos-Fernandez, G.; Augustin, S.; Wehrle, S.; Grossniklaus, U.; Santiago, J. Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. BioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Loubert-Hudon, A.; Mazin, B.D.; Chevalier, É.; Matton, D.P. The ScRALF3 secreted peptide is involved in sporophyte to gametophyte signalling and affects pollen mitosis I. Plant Biol. 2020, 22, 13–20. [Google Scholar] [CrossRef]
- Herger, A.; Dünser, K.; Kleine-Vehn, J.; Ringli, C. Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 2019, 29, R851–R858. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Chakraborti, D.; Basu, D.; Das, S. In search of decoy/guardee to R genes: Deciphering the role of sugars in defense against Fusarium wilt in chickpea. Plant Signal. Behav. 2010, 5, 1081–1087. [Google Scholar] [CrossRef] [Green Version]
- Merino, M.C.; Guidarelli, M.; Negrini, F.; Biase, D.D.; Pession, A.; Baraldi, E. Induced expression of the Fragaria × ananassa rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol. Plant Pathol. 2019, 20, 1252–1263. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; Xie, Z.; Chen, J.; Schnable, P.S.; Walley, J.W.; Yin, Y. Feronia receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef] [Green Version]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 2017, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P.; et al. The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 2014, 9, e92644. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the genome database for rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhang, H.; Sun, L.; Fan, G.; Ye, M.; Jiang, L.; Liu, X.; Ma, K.; Shi, C.; Bao, F.; et al. The genetic architecture of floral traits in the woody plant Prunus mume. Nat. Commun. 2018, 9, 1702. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Firth, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 36, W345–W349. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Duvick, J.; Fu, A.; Muppirala, U.; Sabharwal, M.; Wilkerson, M.D.; Lawrence, C.J.; Lushbough, C.; Brendel, V. PlantGDB: A resource for comparative plant genomics. Nucleic Acids Res. 2008, 36, D959–D965. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y. Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Jambagi, S.; Dunwell, J.M. Global transcriptome analysis and identification of differentially expressed genes after infection of Fragaria vesca with powdery mildew (Podosphaera aphanis). Transcriptomics 2015, 3, 1000106. [Google Scholar] [CrossRef] [Green Version]
- Hollender, C.A.; Kang, C.; Darwish, O.; Geretz, A.; Matthews, B.F.; Slovin, J.; Alkharouf, N.; Liu, Z. Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol. 2014, 165, 1062–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darwish, O.; Shahan, R.; Liu, Z.; Slovin, J.P.; Alkharouf, N.W. Re-annotation of the woodland strawberry (Fragaria vesca) genome. BMC Genom. 2015, 16, 29. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Sevilla, J.F.; Vallarino, J.G.; Osorio, S.; Bombarely, A.; Posé, D.; Merchante, C.; Botella, M.A.; Amaya, I.; Valpuesta, V. Gene expression atlas of fruit ripening and transcriptome assembly from RNA-seq data in octoploid strawberry (Fragaria × ananassa). Sci. Rep. 2017, 7, 13737. [Google Scholar] [CrossRef]
- Jumas-Bilak, E.; Marchandin, H. The phylum synergistetes. Prokaryotes 2014, 931–954. [Google Scholar] [CrossRef]
- Pratas, M.I.; Aguiar, B.; Vieira, J.; Nunes, V.; Teixeira, V.; Fonseca, N.A.; Iezzoni, A.; van Nocker, S.; Vieira, C.P. Inferences on specificity recognition at the Malus × domestica gametophytic self-incompatibility system. Sci. Rep. 2018, 8, 1717. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Ni, W.; Liu, S.; Cai, B.; Xing, H.; Wang, S. Transcriptomics analysis of apple leaves in response to Alternaria alternata apple pathotype infection. Front. Plant Sci. 2017, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- An, N.; Fan, S.; Wang, Y.; Zhang, L.; Gao, C.; Zhang, D.; Han, M. Genome-wide identification, characterization and expression analysis of long non-coding RNAs in different tissues of apple. Gene 2018, 666, 44–57. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Han, M.J.; Jung, K.H.; Yi, G.; Lee, D.Y.; An, G. Rice immature pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. 2006, 47, 1457–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Q.K.; Zhang, W.Z.; Liu, Y.T.; Tao, L.Z.; Liu, H.L. FERONIA regulates auxin-mediated lateral root development and primary root gravitropism. FEBS Lett. 2018, 593, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]



| Symbol | Gene ID | Forward Sequence (5′ to 3′) | Reverse Sequence (5′ to 3′) |
|---|---|---|---|
| Fa-RALF1 | snap_masked-Fvb4-2-processed-gene-95.31-mRNA-1 | CAACCTCCTCATTCTCCTCAC | TGCTGGTGGCTAAGATGC |
| Fa-RALF2 | augustus_masked-Fvb4-2-processed-gene-134.10-mRNA-1 | TCCTCACCCATTTCTCAATC | CCTTCTTCAGTGTCTCGTAGC |
| Fa-RALF5 | snap_masked-Fvb2-1-processed-gene-164.41-mRNA-1 | ACGGGTTGAGCTTTGTTCCT | GGCCAAGATACGCCTGTTGA |
| Fa-RALF8 | snap_masked-Fvb6-1-processed-gene-108.14-mRNA-1 | CATGGAGGCGCAATGTCAAG | AATAATTCTGGCAGTCACCGGA |
| Fa-RALF10 | augustus_masked-Fvb6-1-processed-gene-322.10-mRNA-1 | AGCTGGTGGAGACCTCTCAT | CGGTTATAGGGGTTGGCCTG |
| Fa-RALF12 | augustus_masked-Fvb3-3-processed-gene-31.4-mRNA-1 | ACTTCTTCTGTTTGTCCACCC | GACGACTCTACTGCAGCCAC |
| Fa-RALF13 | augustus_masked-Fvb3-3-processed-gene-31.5-mRNA-1 | CATCAGCTACGGTGCCCTAA | CTGCAACCTCGACGATAGGT |
| Fa-RALF14 | augustus_masked-Fvb1-1-processed-gene-196.6-mRNA-1 | TTCTTACGGTGCGCTCTCAG | GCAGCCTCTGCTGTAAGGAT |
| Fa-RALF18 | augustus_masked-Fvb6-3-processed-gene-378.1-mRNA-1 | TGGATACCCACCACCGTTTC | GCTATAGGGGTTGGCCTGAG |
| Fa-RALF19 | augustus_masked-Fvb1-4-processed-gene-81.5-mRNA-1 | TGACTTCTTCAACTCTACTCCACTA | GCCCTGGCGGAAGGGCAGTCAGGGC |
| Fa-RALF24 | augustus_masked-Fvb1-3-processed-gene-90.5-mRNA-1 | GCTTCTCCAACTCTACTCCACTATC | CGACTCGTCGAGATGGGCCCTGGCG |
| Fa-RALF25 | snap_masked-Fvb6-4-processed-gene-50.20-mRNA-1 | TCTCCGGCTTAGTCTCCGAC | GGCTATAGGGGTTGGCCTGA |
| Fa-RALF29 | augustus_masked-Fvb2-3-processed-gene-83.11-mRNA-1 | CATTGCAGAGTGCATGGCTG | GTAGGGATTAGCCTGTGCCC |
| Fa-RALF31 | snap_masked-Fvb4-3-processed-gene-153.15-mRNA-1 | TAATCTTCTACCTGGGTCTCCTCTT | TTTGAGTGCATTGAGGTCCAGAACT |
| FXARAL32 | maker-Fvb4-3-snap-gene-192.46-mRNA-1 | GCAATGGGTCTTCAGTTCTG | GCAATGGGTCTTCAGTTCTG |
| Fa-RALF33 | augustus_masked-Fvb1-2-processed-gene-103.4-mRNA-1 | ATGACTTCTTCAACTCTACTCCACT | AACTTCACGCTCGCCTCGTCGAGCT |
| Fa-RALF35 | snap_masked-Fvb2-2-processed-gene-47.50-mRNA-1 | GGCAAAGTCCTCTTCCATTATTCTC | CGGGACTTGGCCGGAACAAAGCTCA |
| Fa-RALF39 | snap_masked-Fvb4-4-processed-gene-106.36-mRNA-1 | TCCTCATTCTCCTCACTACTACTCT | GGAAGCCGTGGAATGGGCGGCCAGG |
| Fa-RALF40 | augustus_masked-Fvb4-4-processed-gene-145.10-mRNA-1 | AAGGTCGGGGACTGCATAAC | TCGTGTAATGACCTCACAGCC |
| EF-1α | XM_004307362 | CATGCGCCAGACTGTTGCTGT | GACCGACTCAGAATACTAGTAGC |
| Plant Tissues and Stresses | Genes Retrieved from Expression Sequence Tag (EST) Data |
|---|---|
| Sault | Fv-RALF10, Fv-RALF11, Fv-RALF12 |
| Drought | Fv-RALF10 |
| Heat | Fv-RALF11, Fv-RALF12 |
| Red fruit/Fruit | Fa-RALF5, Fa-RALF29, Fa-RALF35, Md-RALF2, Md-RALF3, Md-RALF 5, Md-RALF6, Md-RALF7, Md-RALF11, Md-RALF12, Md-RALF13, Md-RALF14, Md-RALF15, Md-RALF19, Md-RALF20, Pp-RALF12, Pp-RALF13 |
| Flower | Md-RALF1, Md-RALF9, Md-RALF10, Md-RALF2, Md-RALF5, Md-RALF7, Md-RALF11, Md-RALF12, Md-RALF13, Md-RALF14, Md-RALF 15, Md-RALF 19, Md-RALF 20 |
| Bud | Md-RALF2, Md-RALF3, Md-RALF7, Md-RALF11, Md-RALF13, Md-RALF 15, Md-RALF 19, Md-RALF 20 |
| Young root | Md-RALF3, Md-RALF7, Md-RALF11 |
| Xylem | Md-RALF 6, Md-RALF 7, Md-RALF13, Md-RALF19, Md-RALF20 |
| Young shoot | Md-RALF2, Md-RALF7, Md-RALF13, Md-RALF15, Md-RALF19, Md-RALF 20 |
| Leaf | Md-RALF2, Md-RALF13, Md-RALF15, Md-RALF19, Md-RALF20, Md-RALF, Pp-RALF5 |
| Shoot internodes | Md-RALF 13, Md-RALF 19 |
| Fruit cortex | Md-RALF 13, Md-RALF 19 |
| Fruit core | Md-RALF 13, Md-RALF 19 |
| Phloem | Md-RALF 20 |
| Mesocarp | Pp-RALF6, Pp-RALF8, Pp-RALF13 |
| Venturia inaequalis | Md-RALF5, Md-RALF14 |
| Choristoneura rosaceana | Md-RALF20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Jing, X.; Chen, Y.; Liu, Z.; Xin, Y.; Qiao, Y. The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae). Genes 2020, 11, 174. https://doi.org/10.3390/genes11020174
Zhang H, Jing X, Chen Y, Liu Z, Xin Y, Qiao Y. The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae). Genes. 2020; 11(2):174. https://doi.org/10.3390/genes11020174
Chicago/Turabian StyleZhang, Hong, Xiaotong Jing, Ying Chen, Zhe Liu, Yuting Xin, and Yushan Qiao. 2020. "The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae)" Genes 11, no. 2: 174. https://doi.org/10.3390/genes11020174
APA StyleZhang, H., Jing, X., Chen, Y., Liu, Z., Xin, Y., & Qiao, Y. (2020). The Genome-Wide Analysis of RALF-Like Genes in Strawberry (Wild and Cultivated) and Five Other Plant Species (Rosaceae). Genes, 11(2), 174. https://doi.org/10.3390/genes11020174






