The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA
Abstract
1. Introduction
2. Top1 Binding of Duplex DNA
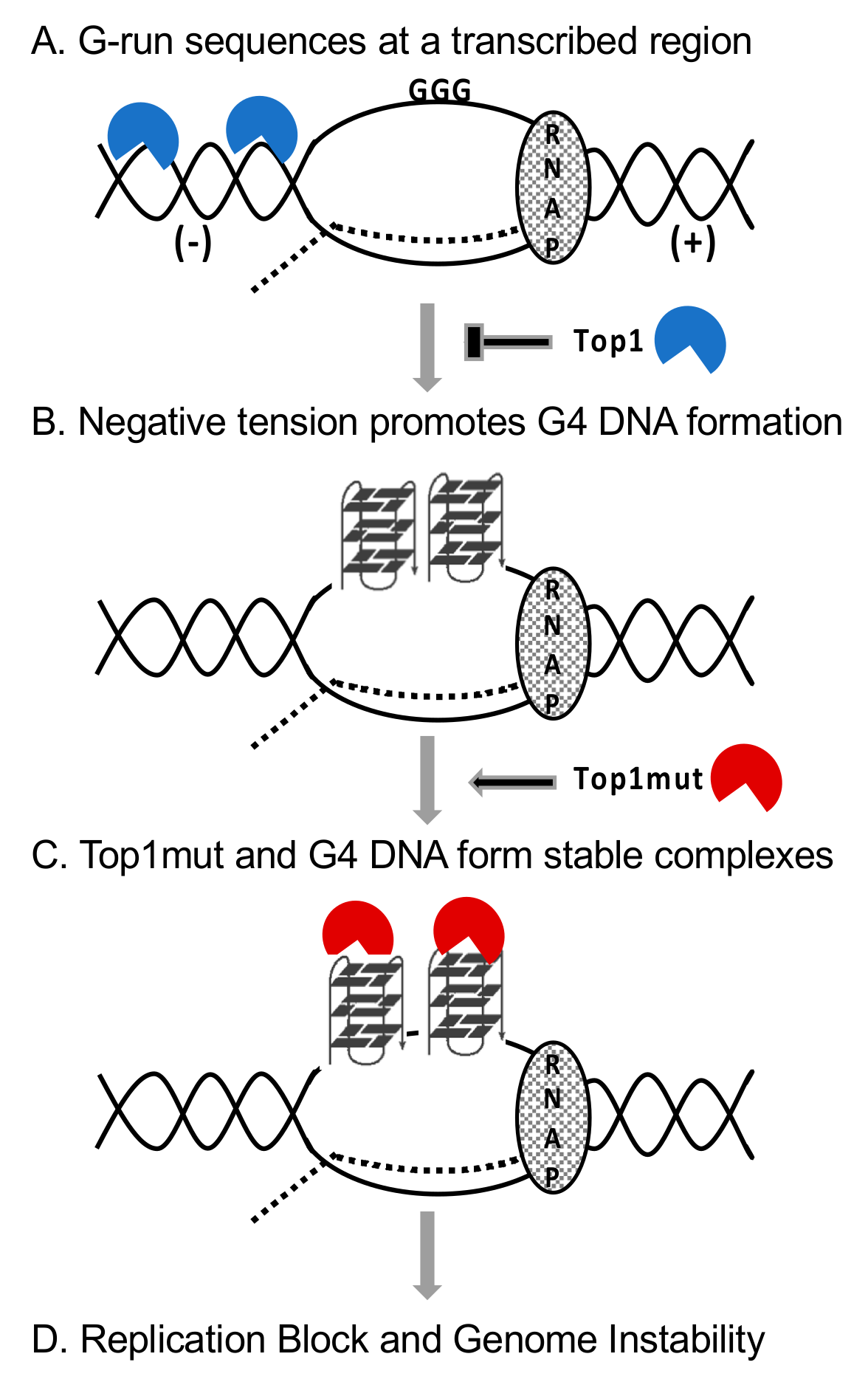
3. The Role of Top1 in Preventing the Formation of G4 DNA and Other DNA Secondary Structures
4. Top1 Binding G4 DNA
5. Functional Consequence of Top1–G4 DNA Binding
5.1. G4 Oligos as Top1 Inhibiting Aptamers
5.2. The Biological Consequence of the Top1 Interaction with G4 DNA
5.3. Interaction between Mutant Top1 and G4 DNA In Vivo
5.4. DNA–Protein Complexes as DNA Replication Barriers
6. Top1 Mutants in Cancer Cells and G4-Induced Genomic Instability
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Sekiguchi, J.; Shuman, S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell 1997, 1, 89–97. [Google Scholar] [CrossRef]
- Kim, N.; Huang, S.Y.; Williams, J.S.; Li, Y.C.; Clark, A.B.; Cho, J.E.; Kunkel, T.A.; Pommier, Y.; Jinks-Robertson, S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 2011, 332, 1561–1564. [Google Scholar] [CrossRef]
- Williams, J.S.; Smith, D.J.; Marjavaara, L.; Lujan, S.A.; Chabes, A.; Kunkel, T.A. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 2013, 49, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.E.; Kim, N.; Li, Y.C.; Jinks-Robertson, S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair 2013. [Google Scholar] [CrossRef] [PubMed]
- Cornelio, D.A.; Sedam, H.N.; Ferrarezi, J.A.; Sampaio, N.M.; Argueso, J.L. Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability. DNA Repair (Amst.) 2017, 52, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.N.; Williams, J.S.; Arana, M.E.; Kunkel, T.A.; Pommier, Y. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 2017, 36, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Sperling, A.S.; Jeong, K.S.; Kitada, T.; Grunstein, M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 12693–12698. [Google Scholar] [CrossRef]
- Pedersen, J.M.; Fredsoe, J.; Roedgaard, M.; Andreasen, L.; Mundbjerg, K.; Kruhoffer, M.; Brinch, M.; Schierup, M.H.; Bjergbaek, L.; Andersen, A.H. DNA Topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 2012, 8, e1003128. [Google Scholar] [CrossRef]
- Baranello, L.; Wojtowicz, D.; Cui, K.; Devaiah, B.N.; Chung, H.J.; Chan-Salis, K.Y.; Guha, R.; Wilson, K.; Zhang, X.; Zhang, H.; et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell 2016, 165, 357–371. [Google Scholar] [CrossRef]
- Davis, L.; Maizels, N. G4 DNA: At risk in the genome. EMBO J. 2011, 30, 3878–3879. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.; Redinbo, M.R.; Qiu, X.; Hol, W.G.; Champoux, J.J. A model for the mechanism of human topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Redinbo, M.R.; Stewart, L.; Kuhn, P.; Champoux, J.J.; Hol, W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 1998, 279, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Madden, K.R.; Stewart, L.; Champoux, J.J. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995, 14, 5399–5409. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Carey, J.F.; Champoux, J.J. Mutational analysis of the preferential binding of human topoisomerase I to supercoiled DNA. FEBS J. 2009, 276, 5906–5919. [Google Scholar] [CrossRef]
- Sekiguchi, J.; Shuman, S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 1994, 22, 5360–5365. [Google Scholar] [CrossRef]
- Been, M.D.; Burgess, R.R.; Champoux, J.J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984, 12, 3097–3114. [Google Scholar] [CrossRef][Green Version]
- Siu, F.M.; Pommier, Y. Sequence selectivity of the cleavage sites induced by topoisomerase I inhibitors: A molecular dynamics study. Nucleic Acids Res. 2013, 41, 10010–10019. [Google Scholar] [CrossRef]
- Aguilera, A.; Gaillard, H. Transcription and recombination: When RNA meets DNA. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria. PLoS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef] [PubMed]
- Manzo, S.G.; Hartono, S.R.; Sanz, L.A.; Marinello, J.; De Biasi, S.; Cossarizza, A.; Capranico, G.; Chedin, F. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, S.M. Discovery of alternative DNA structures: A heroic decade (1979–1989). Front. Biosci. 2008, 13, 1064–1071. [Google Scholar] [CrossRef]
- Irobalieva, R.N.; Fogg, J.M.; Catanese, D.J., Jr.; Sutthibutpong, T.; Chen, M.; Barker, A.K.; Ludtke, S.J.; Harris, S.A.; Schmid, M.F.; Chiu, W.; et al. Structural diversity of supercoiled DNA. Nat. Commun. 2015, 6, 8440. [Google Scholar] [CrossRef] [PubMed]
- Hubert, L., Jr.; Lin, Y.; Dion, V.; Wilson, J.H. Topoisomerase 1 and single-strand break repair modulate transcription-induced CAG repeat contraction in human cells. Mol. Cell. Biol. 2011, 31, 3105–3112. [Google Scholar] [CrossRef]
- Nakatani, R.; Nakamori, M.; Fujimura, H.; Mochizuki, H.; Takahashi, M.P. Large expansion of CTG*CAG repeats is exacerbated by MutSbeta in human cells. Sci. Rep. 2015, 5, 11020. [Google Scholar] [CrossRef]
- Katapadi, V.K.; Nambiar, M.; Raghavan, S.C. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics 2012, 100, 72–80. [Google Scholar] [CrossRef]
- Bacolla, A.; Tainer, J.A.; Vasquez, K.M.; Cooper, D.N. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef]
- Kobayashi, M.; Aida, M.; Nagaoka, H.; Begum, N.A.; Kitawaki, Y.; Nakata, M.; Stanlie, A.; Doi, T.; Kato, L.; Okazaki, I.M.; et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 22375–22380. [Google Scholar] [CrossRef]
- Kobayashi, M.; Sabouri, Z.; Sabouri, S.; Kitawaki, Y.; Pommier, Y.; Abe, T.; Kiyonari, H.; Honjo, T. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc. Natl. Acad. Sci. USA 2011, 108, 19305–19310. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Begum, N.A.; Taniguchi, T.; Taniguchi, H.; Kobayashi, M.; Honjo, T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat. Commun. 2016, 7, 10549. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Harcy, V.; Argueso, J.L.; Dominska, M.; Jinks-Robertson, S.; Kim, N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 2014, 10, e1004839. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yadav, P.; Owiti, N.; Kim, N. The role of topoisomerase I in suppressing genome instability associated with a highly transcribed guanine-rich sequence is not restricted to preventing RNA:DNA hybrid accumulation. Nucleic Acids Res. 2016, 44, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Zyner, K.G.; Mulhearn, D.S.; Adhikari, S.; Martinez Cuesta, S.; Di Antonio, M.; Erard, N.; Hannon, G.J.; Tannahill, D.; Balasubramanian, S. Genetic interactions of G-quadruplexes in humans. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Thiyagarajan, M.M.; Waldman, S.A.; Noe, M.; Kmiec, E.B. Binding characteristics of Ustilago maydis topoisomerase I to DNA containing secondary structures. Eur. J. Biochem. 1998, 255, 347–355. [Google Scholar] [CrossRef]
- Arimondo, P.B.; Riou, J.F.; Mergny, J.L.; Tazi, J.; Sun, J.S.; Garestier, T.; Helene, C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000, 28, 4832–4838. [Google Scholar] [CrossRef]
- Marchand, C.; Pourquier, P.; Laco, G.S.; Jing, N.; Pommier, Y. Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J. Biol. Chem. 2002, 277, 8906–8911. [Google Scholar] [CrossRef]
- Shuai, L.; Deng, M.; Zhang, D.; Zhou, Y.; Zhou, X. Quadruplex-duplex motifs as new topoisomerase I inhibitors. Nucleosides Nucleotides Nucleic Acids 2010, 29, 841–853. [Google Scholar] [CrossRef]
- Fry, M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007, 12, 4336–4351. [Google Scholar] [CrossRef]
- Gao, J.; Zybailov, B.L.; Byrd, A.K.; Griffin, W.C.; Chib, S.; Mackintosh, S.G.; Tackett, A.J.; Raney, K.D. Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem. Commun. 2015, 51, 7242–7244. [Google Scholar] [CrossRef]
- Lopez, C.R.; Singh, S.; Hambarde, S.; Griffin, W.C.; Gao, J.; Chib, S.; Yu, Y.; Ira, G.; Raney, K.D.; Kim, N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Griffin, W.C.; Gao, J.; Byrd, A.K.; Chib, S.; Raney, K.D. A biochemical and biophysical model of G-quadruplex DNA recognition by positive coactivator of transcription 4. J. Biol. Chem. 2017, 292, 9567–9582. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Adrian, M.; Heddi, B.; Serero, A.; Nicolas, A.; Mergny, J.L.; Phan, A.T. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J. Am. Chem. Soc. 2012, 134, 5807–5816. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef] [PubMed]
- Heddi, B.; Phan, A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011, 133, 9824–9833. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Ogloblina, A.M.; Bannikova, V.A.; Khristich, A.N.; Oretskaya, T.S.; Yakubovskaya, M.G.; Dolinnaya, N.G. Parallel G-Quadruplexes Formed by Guanine-Rich Microsatellite Repeats Inhibit Human Topoisomerase I. Biochemistry (Mosc.) 2015, 80, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Megonigal, M.D.; Fertala, J.; Bjornsti, M.A. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 1997, 272, 12801–12808. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Lotito, L.; Russo, A.; Chillemi, G.; Bueno, S.; Cavalieri, D.; Capranico, G. Global transcription regulation by DNA topoisomerase I in exponentially growing Saccharomyces cerevisiae cells: Activation of telomere-proximal genes by TOP1 deletion. J. Mol. Biol. 2008, 377, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Lebel, M.; Spillare, E.A.; Harris, C.C.; Leder, P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 1999, 274, 37795–37799. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Haluska, P., Jr.; Rubin, E.H. A role for the amino terminus of human topoisomerase I. Adv. Enzym. Regul. 1998, 38, 253–262. [Google Scholar] [CrossRef]
- Indig, F.E.; Rybanska, I.; Karmakar, P.; Devulapalli, C.; Fu, H.; Carrier, F.; Bohr, V.A. Nucleolin inhibits G4 oligonucleotide unwinding by Werner helicase. PLoS ONE 2012, 7, e35229. [Google Scholar] [CrossRef]
- Tajrishi, M.M.; Tuteja, R.; Tuteja, N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun. Integr. Biol. 2011, 4, 267–275. [Google Scholar] [CrossRef]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef]
- Hanakahi, L.A.; Sun, H.; Maizels, N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999, 274, 15908–15912. [Google Scholar] [CrossRef]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.; Shaw, P.G.; Kim, M.S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef]
- Weitzmann, M.N.; Woodford, K.J.; Usdin, K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996, 271, 20958–20964. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Reams, C.; Simpson, L.J.; Sale, J.E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell 2010, 40, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Berroyer, A.; Kim, N. The RGG domain of the yeast nucleolin Nsr1 Is required for the genome instability associated with co-transcriptionally formed G4 DNA. BioRxiv 2019, 802876. [Google Scholar] [CrossRef]
- Azevedo, C.; Livermore, T.; Saiardi, A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 2015, 58, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, M.C.; Noguchi, E. Regulation of DNA Replication through Natural Impediments in the Eukaryotic Genome. Genes (Basel) 2017, 8. [Google Scholar] [CrossRef]
- Kamada, K.; Horiuchi, T.; Ohsumi, K.; Shimamoto, N.; Morikawa, K. Structure of a replication-terminator protein complexed with DNA. Nature 1996, 383, 598–603. [Google Scholar] [CrossRef]
- Hill, T.M.; Henson, J.M.; Kuempel, P.L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc. Natl. Acad. Sci. USA 1987, 84, 1754–1758. [Google Scholar] [CrossRef]
- Hill, T.M.; Tecklenburg, M.L.; Pelletier, A.J.; Kuempel, P.L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 1989, 86, 1593–1597. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Sahoo, T.; Bastia, D. The relationship between sequence-specific termination of DNA replication and transcription. EMBO J. 1996, 15, 2530–2539. [Google Scholar] [CrossRef]
- Payne, B.T.; van Knippenberg, I.C.; Bell, H.; Filipe, S.R.; Sherratt, D.J.; McGlynn, P. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res. 2006, 34, 5194–5202. [Google Scholar] [CrossRef] [PubMed]
- Brewer, B.J.; Fangman, W.L. A replication fork barrier at the 3’ end of yeast ribosomal RNA genes. Cell 1988, 55, 637–643. [Google Scholar] [CrossRef]
- Kobayashi, T.; Horiuchi, T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1996, 1, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Woodford, K.J.; Howell, R.M.; Usdin, K. A novel K+-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 1994, 269, 27029–27035. [Google Scholar]
- Berroyer, A.; Alvarado, G.; Larson, E.D. Response of Sulfolobus solfataricus Dpo4 polymerase in vitro to a DNA G-quadruplex. Mutagenesis 2019, 34, 289–297. [Google Scholar] [CrossRef]
- Eddy, S.; Maddukuri, L.; Ketkar, A.; Zafar, M.K.; Henninger, E.E.; Pursell, Z.F.; Eoff, R.L. Evidence for the kinetic partitioning of polymerase activity on G-quadruplex DNA. Biochemistry 2015, 54, 3218–3230. [Google Scholar] [CrossRef]
- Eddy, S.; Tillman, M.; Maddukuri, L.; Ketkar, A.; Zafar, M.K.; Eoff, R.L. Human Translesion Polymerase kappa Exhibits Enhanced Activity and Reduced Fidelity Two Nucleotides from G-Quadruplex DNA. Biochemistry 2016, 55, 5218–5229. [Google Scholar] [CrossRef]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef]
- Ribeyre, C.; Lopes, J.; Boule, J.B.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Boule, J.B.; Lopes, J.; Mingo, K.; Largy, E.; Teulade-Fichou, M.P.; Nicolas, A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010, 38, 4337–4348. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Adrian, M.; Samazan, F.; Heddi, B.; Hamon, F.; Serero, A.; Lopes, J.; Teulade-Fichou, M.P.; Phan, A.T.; Nicolas, A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015, 34, 1718–1734. [Google Scholar] [CrossRef] [PubMed]
- Moruno-Manchon, J.F.; Koellhoffer, E.C.; Gopakumar, J.; Hambarde, S.; Kim, N.; McCullough, L.D.; Tsvetkov, A.S. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY) 2017, 9, 1957–1970. [Google Scholar] [CrossRef]
- Stingele, J.; Schwarz, M.S.; Bloemeke, N.; Wolf, P.G.; Jentsch, S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 2014, 158, 327–338. [Google Scholar] [CrossRef]
- Stingele, J.; Bellelli, R.; Alte, F.; Hewitt, G.; Sarek, G.; Maslen, S.L.; Tsutakawa, S.E.; Borg, A.; Kjaer, S.; Tainer, J.A.; et al. Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Mol. Cell 2016, 64, 688–703. [Google Scholar] [CrossRef]
- Vaz, B.; Popovic, M.; Newman, J.A.; Fielden, J.; Aitkenhead, H.; Halder, S.; Singh, A.N.; Vendrell, I.; Fischer, R.; Torrecilla, I.; et al. Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Mol. Cell 2016, 64, 704–719. [Google Scholar] [CrossRef]
- Maskey, R.S.; Flatten, K.S.; Sieben, C.J.; Peterson, K.L.; Baker, D.J.; Nam, H.J.; Kim, M.S.; Smyrk, T.C.; Kojima, Y.; Machida, Y.; et al. Spartan deficiency causes accumulation of Topoisomerase 1 cleavage complexes and tumorigenesis. Nucleic Acids Res. 2017, 45, 4564–4576. [Google Scholar] [CrossRef]
- Stingele, J.; Bellelli, R.; Boulton, S.J. Mechanisms of DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 563–573. [Google Scholar] [CrossRef]
- Beretta, G.L.; Gatti, L.; Perego, P.; Zaffaroni, N. Camptothecin resistance in cancer: Insights into the molecular mechanisms of a DNA-damaging drug. Curr. Med. Chem. 2013, 20, 1541–1565. [Google Scholar] [CrossRef]
- Zuco, V.; Supino, R.; Favini, E.; Tortoreto, M.; Cincinelli, R.; Croce, A.C.; Bucci, F.; Pisano, C.; Zunino, F. Efficacy of ST1968 (namitecan) on a topotecan-resistant squamous cell carcinoma. Biochem. Pharmacol. 2010, 79, 535–541. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burgess, D.J.; Doles, J.; Zender, L.; Xue, W.; Ma, B.; McCombie, W.R.; Hannon, G.J.; Lowe, S.W.; Hemann, M.T. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 9053–9058. [Google Scholar] [CrossRef]
- Hann, C.L.; Carlberg, A.L.; Bjornsti, M.A. Intragenic suppressors of mutant DNA topoisomerase I-induced lethality diminish enzyme binding of DNA. J. Biol. Chem. 1998, 273, 31519–31527. [Google Scholar] [CrossRef] [PubMed]
- Kubota, N.; Kanzawa, F.; Nishio, K.; Takeda, Y.; Ohmori, T.; Fujiwara, Y.; Terashima, Y.; Saijo, N. Detection of topoisomerase I gene point mutation in CPT-11 resistant lung cancer cell line. Biochem. Biophys. Res. Commun. 1992, 188, 571–577. [Google Scholar] [CrossRef]
- Losasso, C.; Cretaio, E.; Fiorani, P.; D’Annessa, I.; Chillemi, G.; Benedetti, P. A single mutation in the 729 residue modulates human DNA topoisomerase IB DNA binding and drug resistance. Nucleic Acids Res. 2008, 36, 5635–5644. [Google Scholar] [CrossRef]
- Arakawa, Y.; Suzuki, H.; Saito, S.; Yamada, H. Novel missense mutation of the DNA topoisomerase I gene in SN-38-resistant DLD-1 cells. Mol. Cancer Ther. 2006, 5, 502–508. [Google Scholar] [CrossRef]
- Tsurutani, J.; Nitta, T.; Hirashima, T.; Komiya, T.; Uejima, H.; Tada, H.; Syunichi, N.; Tohda, A.; Fukuoka, M.; Nakagawa, K. Point mutations in the topoisomerase I gene in patients with non-small cell lung cancer treated with irinotecan. Lung Cancer 2002, 35, 299–304. [Google Scholar] [CrossRef]
- Mouradov, D.; Sloggett, C.; Jorissen, R.N.; Love, C.G.; Li, S.; Burgess, A.W.; Arango, D.; Strausberg, R.L.; Buchanan, D.; Wormald, S.; et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014, 74, 3238–3247. [Google Scholar] [CrossRef]
- Hurst, C.D.; Alder, O.; Platt, F.M.; Droop, A.; Stead, L.F.; Burns, J.E.; Burghel, G.J.; Jain, S.; Klimczak, L.J.; Lindsay, H.; et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017, 32, 701–715. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.; Sun, X.; Wang, Z.; Zou, X.; Wei, H.; Wang, Z.; Chen, W. Metastatic colorectal cancer and severe hypocalcemia following irinotecan administration in a patient with X-linked agammaglobulinemia: A case report. BMC Med. Genet. 2019, 20, 157. [Google Scholar] [CrossRef] [PubMed]
- Merrouche, Y.; Mugneret, F.; Cahn, J.Y. Secondary acute promyelocytic leukemia following irinotecan and oxaliplatin for advanced colon cancer. Ann. Oncol. 2006, 17, 1025–1026. [Google Scholar] [CrossRef] [PubMed]
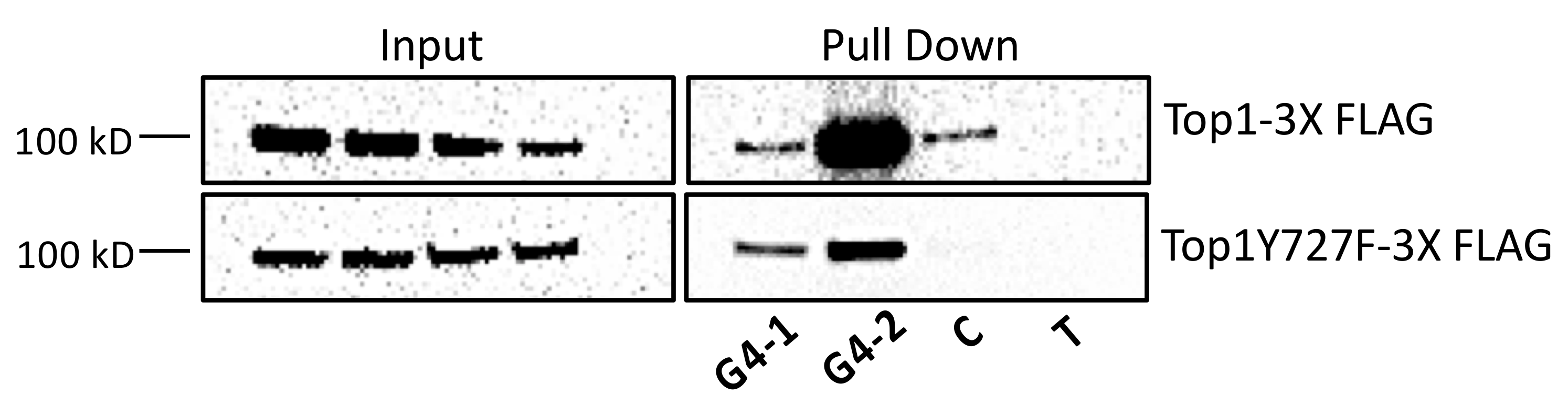
| Oligonucleotide | Sequence |
|---|---|
| G4-1 | 5’ GAGCTGGGGTGAGCTGGGCTGAGCTGGGGTGAGCTGGGCTGAGCT |
| G4-2 | 5’ AGGGCTCTGCCTTGGGGGGGGGGCAGGAAGGGA |
| C | 5’ AGCTCAGCCCAGCTCACCCCAGCTCAGCCCAGCTCACCCCAGCTC |
| T | 5’ GCACGCGTATCTTTTTGGCGCAGGTG |
| Top1 Mutant | Origin | Reference and Mutation ID |
|---|---|---|
| Q713H | breast cancer tissue tumor sample | ICGC(BRCA-US); COSMIC Genomic Mutation ID COSV63696584 |
| I714T | human colorectal cancer cell line | [98]; COSMIC Genomic Mutation ID COSV63693067 |
| R727S | urinary tract cancer tumor sample | [99]; COSMIC Genomic Mutation ID COSV99057653 |
| T729A | irinotecan-resistant human lung cancer cell line | [94] |
| W736C | urinary tract cancer tumor sample | ICGC/GDC(BLCA-US); COSMIC Genomic Mutation ID COSV63695286 |
| W736STOP | non-small cell lung cancer patient treated with irinotecan | [97] |
| E741STOP | urinary tract cancer tumor sample | [100]; COSMIC Genomic Mutation ID COSV63696859 |
| T747P | prostate cancer tumor sample | [101]; COSMIC Genomic Mutation ID COSV63695236 |
| R749W | soft tissue/smooth muscle tumor sample | [100]; COSMIC Genomic Mutation ID COSV63696078 |
| R749Q | large intestine carcinoma sample | ICGC(COAD-US); COSMIC Genomic Mutation ID COSV63696579 |
| A753S | liver cancer tumor sample | ICGC(LICA-CN); COSMIC Genomic Mutation ID COSV63692735 |
| A759T | large intestine carcinoma tumor sample | ICGC(COCA-CN); COSMIC Genomic Mutation ID COSV63694655 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berroyer, A.; Kim, N. The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA. Genes 2020, 11, 193. https://doi.org/10.3390/genes11020193
Berroyer A, Kim N. The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA. Genes. 2020; 11(2):193. https://doi.org/10.3390/genes11020193
Chicago/Turabian StyleBerroyer, Alexandra, and Nayun Kim. 2020. "The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA" Genes 11, no. 2: 193. https://doi.org/10.3390/genes11020193
APA StyleBerroyer, A., & Kim, N. (2020). The Functional Consequences of Eukaryotic Topoisomerase 1 Interaction with G-Quadruplex DNA. Genes, 11(2), 193. https://doi.org/10.3390/genes11020193




