PRL/microRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture and Treatment
2.2. Extraction and Quality Determination of Total RNA
2.3. Triglyceride (TAG) Content and Cholesterol Detection
2.4. Expression Level Detection in Fluorescence Quantitative PCR
2.5. Transfection of miR-183 Mimics and Inhibitor in Primary Cow Mammary Epithelial Cells (CMECs)
2.6. Western Blot Analysis
2.7. Verification of the Dual Luciferase Reporter Gene Targeting IRS1 by miR-183
2.8. Bisulphite Sequencing PCR (BSP) of miR-183
2.9. Statistical Analysis
3. Results
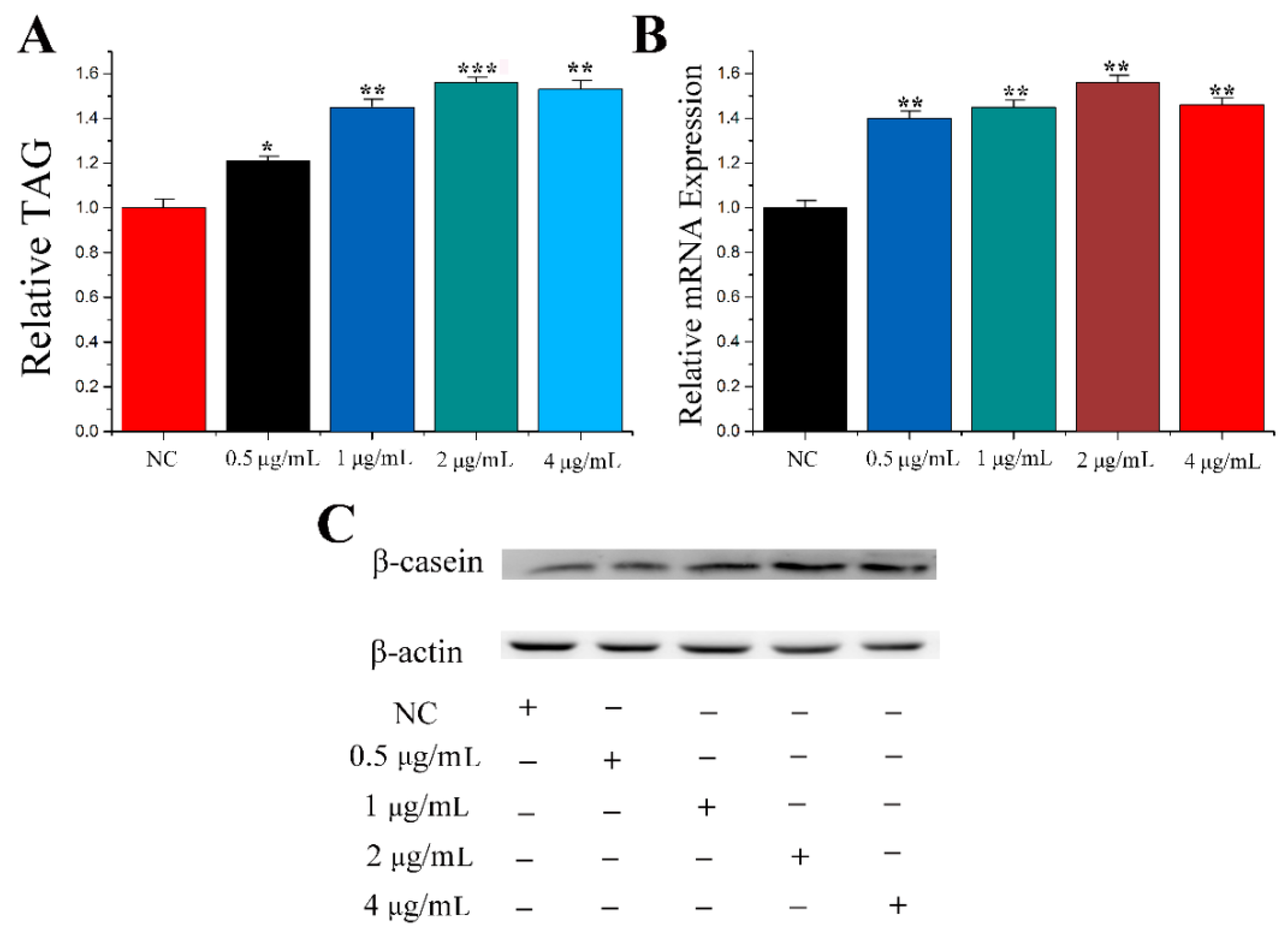
3.1. Screening of the Best PRL Concentration
3.2. Molecular Mechanism of PRL Regulating miR-183 Expression in Cow Mammary Epithelial Cells
3.3. Transfection Efficiency of miRNA and siRNA
3.4. miR-183 Specifically Targets IRS1 in CMECs
3.5. Functions of miR-183 and IRS1 on CMECs
3.6. IRS1 Stimulated the Level of TAG and Increased the Accumulation of Lipid Droplets in CMECs
3.7. siRNA-IRS1 Partially Remedy the Effect of miR-183 on TAG
4. Discussion
4.1. Functions of miR-183 on CMECs
4.2. miR-183 Targets the IRS1 3′-UTR Directly
4.3. PRL Regulates miR-183 Expression by Methylation Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, H.; Yi, R.; Wang, C.; Zhao, P.; Zhang, M.; Xu, S.; Bao, J. Behavior and physiology of two different sow breeds in a farrowing environment during late 35-day lactation. PLoS ONE 2018, 13, e0197152. [Google Scholar] [CrossRef] [Green Version]
- Pillay, J.; Davis, T.J. Physiology, Lactation; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Sriraman, N.K. The Nuts and Bolts of Breastfeeding: Anatomy and Physiology of Lactation. Curr. Probl. Pediatr. Adolesc. Health Care 2017, 47, 305–310. [Google Scholar] [CrossRef]
- Invernizzi, G.; Naeem, A.; Loor, J.J. Short communication: Endoplasmic reticulum stress gene network expression in bovine mammary tissue during the lactation cycle. J. Dairy Sci. 2012, 95, 2562–2566. [Google Scholar] [CrossRef] [Green Version]
- Weng, X.; Monteiro, A.P.A.; Guo, J.; Ahmed, B.M.S.; Bernard, J.K.; Tomlinson, D.J.J.; DeFrain, M.; DahI, G.E.; Tao, S. Short communication: Repeated mammary tissue collections during lactation do not alter subsequent milk yield or composition. J. Dairy Sci. 2017, 100, 8422–8425. [Google Scholar] [CrossRef] [Green Version]
- Horsham, J.L.; Ganda, C.; Kalinowski, F.C.; Brown, R.A.M.; Epis, M.R.; Leedman, P.J. MicroRNA-7: A miRNA with expanding roles in development and disease. Int. J. Biochem. Cell Biol. 2015, 69, 215–224. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, X.; Zhou, Q.; Wang, H.; Qian, Y.; Tian, W.; Ma, D.; Li, X. microRNA expression profiling of heart tissue during fetal development. Int. J. Mol. Med. 2014, 33, 1250–1260. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, L.; Xing, L.P.; Chen, D. MicroRNA-204 Regulates Runx2 Protein Expression and Mesenchymal Progenitor Cell Differentiation. Stem Cells 2010, 28, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.M.; Zhang, H.Y.; Wang, Y.X.; Wang, W.B. MicroRNA-137 is downregulated in human osteosarcoma and regulates cell proliferation and migration through targeting FXYD6. J. Drug Target. 2016, 24, 102–110. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Suárez, Y.; Rayner, K.J.; Moore, K.J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 2011, 22, 86–92. [Google Scholar] [CrossRef]
- Lynn, F.C. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol. Metab. 2009, 20, 452–459. [Google Scholar] [CrossRef]
- Dahlmans, D.; Houzelle, A.; Andreux, P.; Wang, X.; Jorgensen, J.A.; Moullan, N.; Daemen, S.; Kersten, S.; Auwerx, L.; Hoeks, J. MicroRNA-382 silencing induces a mitonuclear protein imbalance and activates the mitochondrial unfolded protein response in muscle cells. J. Cell. Physiol. 2019, 234, 6601–6610. [Google Scholar] [CrossRef] [Green Version]
- Bu, D.; Bionaz, M.; Wang, M.; Nan, X.; Ma, L.; Wang, J. Transcriptome difference and potential crosstalk between liver and mammary tissue in mid-lactation primiparous dairy cows. PLoS ONE 2017, 12, e0173082. [Google Scholar] [CrossRef]
- Jiang, M.; Lee, J.N.; Bionaz, M.; Deng, X.Y.; Wang, Y. Evaluation of suitable internal control genes for RT-qPCR in yak mammary tissue during the lactation cycle. PLoS ONE 2016, 11, e0147705. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eleswarapu, S.; Jiang, H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007, 581, 981–988. [Google Scholar] [CrossRef] [Green Version]
- Avril-Sassen, S.; Goldstein, L.D.; Stingl, J.; Blenkiron, C.; Le Quesne, J.; Spiteri, I.; Karagavriilidou, K.; Watson, C.J.; Tavare, S.; Miska, E.A.; et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genom. 2009, 10, 548. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.Y.; Chen, C.; Xu, X.; Lu, Q. miR-29a promotes pathological cardiac hypertrophy by targeting the PTEN/AKT/mTOR signalling pathway and suppressing autophagy. Acta Physiol. 2019, 227, e13323. [Google Scholar] [CrossRef]
- Xu, H.F.; Luo, J.; Ma, G.Z.; Zhang, X.Y.; Yao, D.W.; Li, M.; Loor, J.J. Acyl-CoA synthetase short-chain family member 2 (ACSS2) is regulated by SREBP-1 and plays a role in fatty acid synthesis in caprine mammary epithelial cells. J. Cell. Physiol. 2018, 233, 1005–1016. [Google Scholar] [CrossRef]
- Wang, H.; Shi, H.P.; Luo, J.; Yi, R.Q.; Yao, D.W.; Zhang, X.Y.; Ma, G.Z.; Loor, J.J. MiR-145 regulates lipogenesis in goat mammary cells via targeting INSIG1 and epigenetic regulation of lipid-related genes. J. Cell. Physiol. 2017, 232, 1030–1040. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, J.; Zhang, C.; Ma, Y.; Sun, S.; Zhang, T.; Loor, J.J. Mechanism of prolactin inhibition of miR-135b via methylation in goat mammary epithelial cells. J. Cell. Physiol. 2018, 233, 651–662. [Google Scholar] [CrossRef]
- Hansen, H.O.; Grunnet, I.; Knudsen, J. Triacylglycerol synthesis in goat mammary gland. The effect of ATP, Mg2+ and glycerol 3-phosphate on the esterification of fatty acids synthesized de novo. Biochem. J. 1984, 220, 513–519. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [Green Version]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine mammary protein synthesis during thelactation cycle. Bioinform. Biol. Insights 2011, 5, 83–98. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef]
- Chen, Z.; Chu, S.; Wang, X.; Fan, Y.; Zhan, T.; Arbab, A.A.I.; Li, M.; Zhang, H.; Mao, Y.; Loor, J.J.; et al. MicroRNA-106b regulates milk fat metabolism via ATP binding cassette subfamily A member 1 ( ABCA1) in bovine mammary epithelial cells. J. Agric. Food Chem. 2019, 67, 3981–3990. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, H.; Sun, S.; Xu, H.; Cao, D.; Luo, J. MicroRNA-181b suppresses TAG via target IRS2 and regulating multiple genes in the Hippo pathway. Exp. Cell Res. 2016, 348, 66–74. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, H.; Sun, S.; Luo, J.; Zhang, W.; Hou, Y.; Loor, J.J. MiR-183 regulates milk fat metabolism via MST1 in goat mammary epithelial cells. Gene 2018, 646, 12–19. [Google Scholar] [CrossRef]
- Chen, C.; Deng, B.; Qiao, M.; Zheng, R.; Chai, J.; Ding, Y.; Peng, J.; Jiang, S. Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs. PLoS ONE 2012, 7, e31426. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Xiang, H.; Peng, Y.L.; Peng, J.; Jiang, S.W. Mature miR-183, negatively regulated by transcription factor GATA3, promotes 3T3-L1 adipogenesis through inhibition of the canonical Wnt/β-catenin signaling pathway by targeting LRP6. Cell. Signal. 2014, 26, 1155–1165. [Google Scholar] [CrossRef]
- Chirita-Emandi, A.; Munteanu, D.; Andreescu, N.; Tutac, P.; Paul, C.; Velea, I.P.; Pusztai, A.M.; Hlistun, V.; Boiciuc, C.; Sacara, V.; et al. No clinical utility of common polymorphisms in IGF1, IRS1, GCKR, PPARG, GCK1 and KCTD1 genes previously associated with insulin resistance in overweight children from Romania and Moldova. J. Pediatr. Endocrinol. Metab. 2019, 32, 33–39. [Google Scholar] [CrossRef]
- Fettiplace, M.R.; Kowal, K.; Ripper, R.; Young, A.; Lis, K.; Rubinstein, I.; Bonini, M.; Minshall, R.; Weinberg, G. Insulin signaling in bupivacaine-induced cardiac toxicity: Sensitization during recovery and potentiation by lipid emulsion. Anesthesiology 2016, 124, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yang, C.; Lin, G.; Chen, Y.; Miao, S.; Liu, B.; Zhao, C. Antidiabetic potential of green seaweed enteromorpha prolifera flavonoids regulating insulin signaling pathway and gut microbiota in type 2 diabetic mice. J. Food Sci. 2019, 84, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazibuko-Mbeje, S.E.; Dludla, P.V.; Roux, C.; Johnson, R.; Ghoor, S.; Joubert, E.; Louw, J.; Opoku, A.R.; Muller, C.J.F. Aspalathin-enriched green rooibos extract reduces hepatic insulin resistance by modulating PI3K/AKT and AMPK pathways. Int. J. Mol. Sci. 2019, 20, 633. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Reddy, J.K. Transcription coactivators for peroxisome proliferator-activated receptors. Biochim. Biophys. Acta 2007, 1771, 936–951. [Google Scholar] [CrossRef] [PubMed]
- Bouzakri, K.; Zachrisson, A.; Al-Khalili, L.; Zhang, B.B.; Koistinen, H.A.; Krook, A.; Zierath, J.R. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006, 4, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Previs, S.F.; Withers, D.J.; Ren, J.M.; White, M.F.; Shulman, G.I. Contrasting effects of IRS-1 versus IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J. Biol. Chem. 2000, 275, 38990–38994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clement, T.; Salone, V.; Rederstorff, M. Dual Luciferase Gene Reporter Assays to Study miRNA Function. In Small Non-Coding RNAs; Rederstorff, M., Ed.; Humana Press: New York, NY, USA, 2015. [Google Scholar]
- Beaudry, K.L.; Parsons, C.L.; Ellis, S.E.; Akers, R.M. Localization and quantitation of macrophages, mast cells, and eosinophils in the developing bovine mammary gland. J. Dairy Sci. 2016, 99, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Brady, N.J.; Chuntova, P.; Schwertfeger, K.L. Macrophages: Regulators of the inflammatory microenvironment during mammary gland development and breast cancer. Mediat. Inflamm. 2016, 2016, 4549676. [Google Scholar] [CrossRef] [Green Version]
- Gangisetty, O.; Wynne, O.; Jabbar, S.; Nasello, C.; Sarkar, D.K. Fetal alcohol exposure reduces dopamine receptor D2 and increases pituitary weight and prolactin production via epigenetic mechanisms. PLoS ONE 2015, 10, e0140699. [Google Scholar] [CrossRef]
- Singh, K.; Erdman, R.A.; Swanson, K.M.; Molenaar, A.J.; Maqbool, N.J.; Wheeler, T.T.; Arias, J.A.; Quinn-Walsh, E.C.; Stelwagen, K. Epigenetic regulation of milk production in dairy cows. J. Mammary Gland Biol. Neoplasia 2010, 15, 101–112. [Google Scholar] [CrossRef]
- Babion, I.; De Strooper, L.M.A.; Luttmer, R.; Bleeker, M.C.G.; Meijer, C.; Heideman, D.A.M.; Wilting, S.M.; Steenbergen, R.D.M. Complementarity between miRNA expression analysis and DNA methylation analysis in hrHPV-positive cervical scrapes for the detection of cervical disease. Epigenetics 2019, 14, 558–567. [Google Scholar] [CrossRef] [Green Version]
- Merkerova, M.D.; Remesova, H.; Krejcik, Z.; Loudova, N.; Hrustincova, A.; Szikszai, K.; Cermak, J.; Jonasova, A.; Belickova, M. Relationship between Altered miRNA Expression and DNA Methylation of the DLK1-DIO3 Region in Azacitidine-Treated Patients with Myelodysplastic Syndromes and Acute Myeloid Leukemia with Myelodysplasia-Related Changes. Cells 2018, 7, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, P.; Yuan, Y.; Zhang, M.; Sun, Y.; Wei, C.; Xie, X.; Zhang, Y.; Wang, S.; Chen, Z.; Wang, X. PRL/microRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells. Genes 2020, 11, 196. https://doi.org/10.3390/genes11020196
Jiao P, Yuan Y, Zhang M, Sun Y, Wei C, Xie X, Zhang Y, Wang S, Chen Z, Wang X. PRL/microRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells. Genes. 2020; 11(2):196. https://doi.org/10.3390/genes11020196
Chicago/Turabian StyleJiao, Peixin, Yuan Yuan, Meimei Zhang, Youran Sun, Chuanzi Wei, Xiaolai Xie, Yonggen Zhang, Sutian Wang, Zhi Chen, and Xiaolong Wang. 2020. "PRL/microRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells" Genes 11, no. 2: 196. https://doi.org/10.3390/genes11020196
APA StyleJiao, P., Yuan, Y., Zhang, M., Sun, Y., Wei, C., Xie, X., Zhang, Y., Wang, S., Chen, Z., & Wang, X. (2020). PRL/microRNA-183/IRS1 Pathway Regulates Milk Fat Metabolism in Cow Mammary Epithelial Cells. Genes, 11(2), 196. https://doi.org/10.3390/genes11020196





